Faith I. Ebhodaghe, Irma Sanchez-Vargas, Clement Isaac, Brian D. Foy & Elizabeth Hemming-Schroeder
Abstract
Background
When integrated with insecticide-treated bed nets, larval control of Anopheles mosquitoes could fast-track reductions in the incidence of human malaria. However, larval control interventions may deliver suboptimal outcomes where the preferred breeding places of mosquito vectors are not well known. This study investigated the breeding habitat choices of Anopheles mosquitoes in southern Nigeria. The objective was to identify priority sites for mosquito larval management in selected urban and periurban locations where malaria remains a public health burden.
Methods
Mosquito larvae were collected in urban and periurban water bodies during the wet-dry season interface in Edo, Delta, and Anambra States. Field-collected larvae were identified based on PCR gel-electrophoresis and amplicon sequencing. In contrast, the associations between Anopheles larvae and the properties and locations of water bodies were assessed using various statistical methods.
Results
Mosquito breeding sites were either man-made (72.09%) or natural (27.91%) and mostly drainages (48.84%) and puddles (25.58%). Anopheles larvae occurred in drainages, puddles, stream margins, and a concrete well, and were absent in drums, buckets, car tyres, and a water-holding iron pan, all of which contained culicine larvae. Wild-caught Anopheles larvae comprised Anopheles coluzzii (80.51%), Anopheles gambiae sensu stricto (s.s.) (11.54%), and Anopheles arabiensis (7.95%); a species-specific PCR confirmed the absence of the invasive urban malaria vector Anopheles stephensi among field-collected larvae. Anopheles arabiensis, An. coluzzii, and An. gambiae s.s. displayed preferences for turbid, lowland, and partially sunlit water bodies. Furthermore, An. arabiensis preferred breeding sites located outside 500 m of households, whereas An. gambiae s.s. and An. coluzzii had increased detection odds in sites within 500 m of households. Anopheles gambiae s.s. and An. coluzzii were also more likely to be present in natural water bodies; meanwhile 96.77% of An. arabiensis were in man-made water bodies. Intraspecific genetic variations were little in the dominant vector An. coluzzii, while breeding habitat choices of populations made no statistically significant contributions to these variations.
Conclusion
Sibling malaria vectors in the An. gambiae complex displays divergent preferences for aquatic breeding habitats in southern Nigeria. The findings are relevant for planning targeted larval control of An. coluzzii whose increasing evolutionary adaptations to urban ecologies are driving the proliferation of the mosquito, and An. arabiensis whose adults typically evade the effects of treated bed nets due to exophilic tendencies.
Background
Malaria remains a major public health challenge, with a disproportionately high burden of infections in Africa; ~ 95% of infection cases and ~ 96% of associated deaths are reported in the continent annually 1. Anopheles gambiae sensu lato (s.l.) are the primary vectors of malaria in sub-Saharan Africa. These mosquitoes breed in clean and natural aquatic environments in the form of small sunlit water collections. However, deviations from this traditionally and widely known choice of breeding habitats by An. gambiae s.l. have been observed 2,3, but these deviations remain understudied in countries in the West African sub-region. Furthermore, An. gambiae s.l. is a complex of mosquitoes comprising more than eight sibling species 4. The anthropophilic and indoor-biting Afrotropical vectors An. coluzzii and An. gambiae sensu stricto (s.s.) —previously known as M and S molecular forms of An. gambiae, respectively— are sibling members of this complex and contribute to the high risk of malaria in Nigeria and neighbouring West and Central African countries 5.
Different mosquito larval surveys in West and Central Africa observed variations in the breeding habitat choices of An. coluzzii and An. gambiae s.s. (reviewed in 6). In Burkina Faso, An. coluzzii co-existed with An. gambiae s.s. but preferred to breed in large, permanent, and vegetation-dense habitats (rice paddies), whereas An. gambiae s.s. preferred temporary puddles 7. Additional differences in the ecologies of An. coluzzii and An. gambiae s.s. have been described in Mali 8 and Cameroon 9 among countries in the West and Central African sub-regions. Furthermore, evidence has emerged supporting the hypothesis that contrasting responses of larvae to breeding habitat conditions formed the basis for ecological speciation of An. coluzzii and An. gambiae s.s. 10. However, underlying ecological factors underpin oviposition site preferences of gravid females and the water properties that mediate segregation of the breeding habitats of An. coluzzii and An. gambiae s.s., are less well known. Addressing this knowledge gap, especially where An. coluzzii and An. gambiae s.s. are sympatric, which is essential to reliably predict the spatial distribution of larvae of the vector species and identify potential sites for targeted and species-specific mosquito larval control interventions.
Mosquito larval control interventions are effective for the control of malaria vectors. They also simultaneously target Anopheles and culicine disease vectors where these mosquitoes co-breed in water bodies 11. Larval control interventions leverage biopesticides or predators to reduce the number of immature mosquitoes in aquatic environments and, where possible, may eliminate water bodies providing breeding places for mosquitoes 12. However, mosquito larval control requires a clear understanding of the breeding habitats of target vectors to accurately select priority sites for interventions. Meanwhile, the World Health Organization 12 recommends mosquito larval control as a suitable method for supplementing pyrethroid-treated bed nets. This is because larval control reduces the abundance of pyrethroid-resistant mosquitoes, as well as outdoor-biting malaria vectors (e.g. Anopheles arabiensis) whose adults are typically outside the reach of pyrethroid-treated bed nets currently widely used in sub-Saharan African countries to control indoor-biting vectors.
Nigeria in West Africa reports the highest malaria disease burden worldwide, with > 25% of the global incidence of infections occurring in the country 1. There are few reports on malaria spread by outdoor-biting mosquito species in Nigeria 13. However, the typically indoor-biting vectors An. coluzzii and An. gambiae s.s. have been found to feed on humans in outdoor locations 14. Some investigators attribute this to a behavioural response by vectors to the protracted use of pyrethroid-treated bed nets 15,16. Long-term adoption of treated bed nets has increased the frequencies of pyrethroid-resistant vectors in wild mosquito populations in southern Nigeria 17,18, thus further compromising the efficacy of treated bed nets for malaria vector control. Although alternative intervention strategies, for example, mosquito larval control, are available for the management of malaria vectors, these strategies have received limited attention in southern Nigeria mainly due to the relatively low economic costs of using pyrethroid-treated bed nets.
Mosquito larval control interventions to manage pyrethroid-resistant An. coluzzii and An. gambiae s.s. could contribute to malaria risk reduction in southern Nigeria. These interventions could also assist in alleviating the epidemiologic burden of outdoor-biting Anopheles vectors that may be locally endemic but evade the effects of pyrethroid-treated bed nets. This study assessed the species diversity of Anopheles malaria vectors in selected urban and periurban areas in southern Nigeria. It further assessed vectors for differences in the choice of breeding habitats. Water bodies were surveyed for the presence and abundance of larvae and their physicochemical properties characterised. Findings from the study add to current knowledge on the larval ecology of An. gambiae s.l. malaria vectors in Africa and provide relevant data for community-led and species-specific larval control interventions in urban and periurban settings in southern Nigeria where malaria risks are currently high and escalating.
Methods
Study area
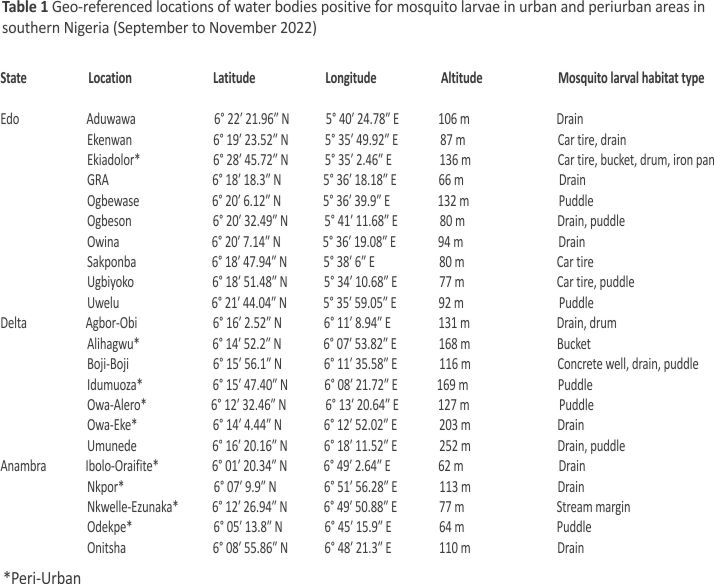
Mosquito larval samplings were done in southern Nigeria with sites spread over a geographical distance of ≈200 km extending from Edo State (6° 17ʹ 1.341″ N, 5° 33ʹ 59.061″ E) to Delta State (6° 12ʹ 6.523″ N, 6° 10ʹ 47.316″ E) and Anambra State (6° 7ʹ 9.12″ N, 6° 47ʹ 15.792″ E). The human population size in these three neighbouring States is ≈16 million 19. According to the WHO Africa 19, household parasite screening surveys in 2021 based on the Rapid Diagnostic Technique (RDT) in children under 5 years of age indicated malaria infection rates of 30.2%, 18.9%, and 20.2% in Edo, Delta, and Anambra, respectively. Human exposure to infections is high in these areas during the wet season (May to mid-October), compared to the dry season (mid-October to April) when the numbers of vector breeding habitats are fewer. The average annual rainfall amount and temperature in southern Nigeria are 2500 mm and 27 ℃, respectively, while vegetation is typically rainforest with extensive networks of freshwater swamps, and sparse and scattered woodlands 20.
Mosquito larvae sampling
Larval samplings were done during the late wet season and the early dry season from September to November 2022. To collect mosquito larvae in urban and periurban water bodies, a standard dipper (300 ml, John W. Hock’s Company, Gainesville, Florida, USA) was lowered towards a water body and carefully but quickly applied to scoop the water surface. Where present in a water sample, mosquito larvae were morphologically identified as either Anopheles or culicine, counted, and stored in alcohol within small, labelled vials. To estimate average larval abundance, the overall number of larvae collected in a water body was divided by the total number of dips made in the same water body.
Characterization of water bodies
Water bodies within 500 m of households were considered ‘close’ while those outside 500 m were considered ‘far’. A water body was ‘turbid’ if it was difficult to see through a water sample and ‘non-turbid’ if otherwise. To determine the depth of mosquito larval sites, a straight pole was inserted in a vertical position into a water body until the pole reached the bottom. Careful notice was made of the watermark on the pole after it had been removed from the water, while a graduated tape was used to measure the pole from the watermark down to the tip that touched the bottom of the water body. Water bodies were considered ‘deep’ if they had a depth of above 20 cm and ‘shallow’ if depths were below 20 cm. A handheld GPS device (Garmin etrex 10) was used to record geographic coordinates and altitude of sampling sites. Water samples were assessed for ‘temperature’, ‘pH’, and ‘salinity’ at each site using a calibrated multi-parametric device (Hanna instrument GroLine Meter) powered by lithium batteries. Measurements of altitude (metres above sea level), temperature (℃), pH, and salinity (parts per million) were considered high if they exceeded the 65th percentile values of their respective distributions, otherwise, they were low. The 65th percentile values for altitude, temperature, pH, and salinity were 136 m (62 m to 316 m, SD: ± 57.46), 30.03 °C (22.7 ℃ to 36.1 ℃, SD: ± 2.76), 7.74 (6.61 to 9.04, SD: ± 0.76), and 140 ppm (0 ppm to 410 ppm, SD: ± 101.79), respectively. Additional data collected at mosquito larval sites were the area (residential or industrial), site location (urban or periurban), habitat type (man-made or natural), vegetation presence (yes or no), presence of debris (yes or no), and water exposure to sunlight (partial or complete).
Molecular identification of Anopheles larvae
Genomic DNA was extracted from each Anopheles larva following the Chelex protocol described by Musapa et al. 21. The Polymerase Chain Reaction (PCR) gel electrophoresis method was used to identify Anopheles larvae and to differentiate species of An. gambiae s.l. by targeting the S200 X6.1 insertion polymorphism present in An. coluzzii but absent in An. gambiae s.s., adopting the primers described by Santolamazza et al. 22 (Fwd: TCGCCTTAGACCTTGCGTTA and Rev: CGCTTCAAGAATTCGAG ATAC). PCR was conducted in 12.5 µl reaction volume containing 1 µl template DNA, 0.25 µl (10 µM) of each primer, 4.75 µl nuclease-free water, and 6.25 µl OneTaq® Quick-Load® 2X Master Mix (New England Biolabs).
PCR for Anopheles DNA amplification targeting the S200 X6.1 gene was carried out on a Thermal cycler (Eppendorf Mastercycler nexus gradient) at 94 ℃ for 30 s; 30 cycles of denaturation at 94 ℃ for 15 s, 54 ℃ for 30 s and 68 ℃ for 1 min, and a final extension at 68 ℃ for 5 min. Anopheles larvae were identified based on a base-pair size of ~ 479 An. coluzzii (M form) and ~ 249 for An. gambiae s.s. (S form). The species An. gambiae s.s. has similar base-pair sizes with its siblings namely An. arabiensis, An. melas, and An. quadriannulatus.
DNA amplifications of the ITS2 gene were carried out to identify samples that failed to amplify the S200 X6.1 gene using the primers ITS2A (Fwd: TGTGAACTGCAGG ACACAT) and ITS2B (Rev: TATGCTTAAATTCAGGGGGT), with reaction volume as described for the S200 X6.1 PCR above. PCR adopted reaction conditions similar to those described previously 23. PCR cycling was carried out on a Thermal cycler (Eppendorf Master- cycler nexus gradient) at 95 ℃ for 3 min; 35 cycles of denaturation at 94 ℃ for 30 s, 55 ℃ for 30 s, and 72 ℃ for 45 s, and a final extension at 72 ℃ for 6 min. Band size for Anopheles using the ITS2 gene marker was ~ 750 bp.
In an attempt to identify samples whose DNA failed to amplify in S200 X6.1-PCR and ITS2-PCR, an additional set of primers was used in endpoint-PCR namely, St-F (CGTATCTTTCCTCGC ATCCA) targeting a region of the ITS2 gene specific to An. stephensi and the universal primers U5.8S-F (ATCACTC GGCTCATGGATCG) and UD2-R (GCAC TATCAAGCAACACGACT) 24. PCR was carried out in 12.5 µl reaction volume containing 1 µl template DNA, 0.25 µl (10 µM) of each of the primers St-F and U5.8S-F and 0.4 µl (10 µM) of the primer UD2-R, 4.4 µl nuclease-free water, and 6.20 µl OneTaq® Quick-Load® 2X Master Mix (New England Biolabs). Cycling was carried out on a Thermal cycler (Eppendorf Mastercycler nexus gradient) at 95 ℃ for 30 s; 30 cycles of denaturation at 95 ℃ for 30 s, 55 ℃ for 30 s, and 68 ℃ for 45 s, and a final extension at 68 ℃ for 7 min. Band size for An. stephensi was ~438 bp with an internal control band of ~ 900 bp.
Nucleotide sequencing and phylogenetic analysis
The species identities of Anopheles were confirmed by unidirectional sequencing at Azenta Life Sciences, Colorado State University, USA. Sequencing was carried out on cleaned PCR-products (Exo-CIP™, New England Biolabs) using the S200 X6.1 primer sequence TCGC CTTAGACCTTGCGTTA 22 and the ITS2B primer sequence TATGCTTAAATTCAGGGGGT 23. DNA sequences were visually inspected for quality in the BioEdit software 25. Good-quality sequences were queried in BLAST analyses on the NCBI website 26. Notes were taken of the sequence identities of query sequences compared to sequences of closest match in the GenBank. Clustal Omega 27 was used to align the study and GenBank sequences, while the Smart Model Selection criterion in PhyML 28 was used to infer the best model of sequence evolution (Hasegawa-Kishino-Yano, HKY) 29. Maximum-likelihood phylogenetic trees were constructed in the software Molecular Evolution and Genetic Analysis MEGA-X 30, and the nodal support values of trees were estimated from 1000 bootstrap replications. Finally, genetic analyses to determine haplotype diversity (Hd) and polymorphic site number of An. coluzzii populations were carried out in DnaSP 31, while haplotype analyses were carried out using Median-Joining (MJ) networks 32. MJ networks were constructed in the PopART software 33 to visualize relationships between populations of An. coluzzii larvae are collected from water bodies in different geographical locations and have different properties.
Data analyses
The numbers of: (i) water body sites with mosquito larvae, (ii) geo-referenced locations surveyed, and (iii) mosquito larval collections were expressed in percentage frequencies with 95% confidence intervals (CI). Differences between percentage frequencies were assessed using a two-proportion Z-test.
To determine their relative importance, predictors of larvae presence in aquatic environments were ranked based on Random Forest (RF) classification analyses. RF analyses were based on 10,000 iterations (ntrees) with 4 variables randomly selected at each split (mtry = √q where q = the total number of variables (= 14)). The R functions ‘importance ()’ and ‘varImpPlot ()’ both embedded in the randomForest package version 4.7–1.1 34 were used to generate Mean Decrease Gini (MDG) scores and variable importance plots, respectively. Variables with higher MDG scores were more important predictors of Anopheles larval presence in water bodies.
Generalized Linear Models (GLM) were used to assess associations between larvae and categorical predictor variables. Generalized Linear Models were fitted assuming a binomial distribution if the response variable was binomial (“larvae_yes” or “larvae_no”), a negative binomial distribution if the response variable was count (number of larvae per dip), and a quasi-binomial distribution if the response variable was proportion (the number of larvae identified for a species divided by the total number of larvae analysed). A GLM assuming a binomial distribution was also used to assess the associations between An. coluzzii haplotypes and populations.
Variations in average larvae abundance were assessed using the Mann–Whitney U test. Correlations between larvae abundance and the following continuous variables (i) altitude, (ii) temperature, (iii) salinity, (iv) culicine abundance, and (v) pH were assessed in Spearman correlation tests and Principal Components Analysis (PCA). A Multiple Correspondence Analysis (MCA) was carried out to visually explore associations between larvae and water properties, as well as between larvae and the location of mosquito breeding sites. PCA and MCA biplots were designed using the R packages ‘FactorMineR‘ and ‘factorextra’ 35.
Multivariate Regression Models were fitted to account for possible confounding effects of variables. Predictor variables were selected for multivariate regression if they had P < 0.05 in univariate models. Furthermore, the backward elimination method was adopted to select predictor variables for the final multivariate model assuming a binomial distribution for binomial response variables, negative binomial distribution for count response variables, and quasi-binomial distribution for proportion response variables. Fisher’s Exact test was used to assess predictor variables for association. Associated predictor variables were mutually exclusive in the final model. Multivariate analyses were followed by pairwise comparisons with Tukey’s adjustment using the function ‘emmeans’ embedded in the ‘emmeans’ package 36. All analyses were carried out in the R Statistical environment 37 while P values were set at an alpha of 0.05.
Results
Mosquito breeding habitats
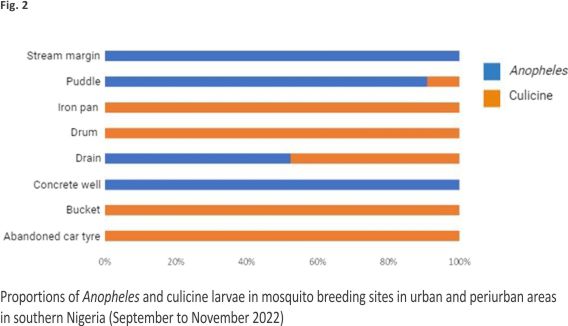
A total of 43 water bodies in aquatic environments surveyed contained mosquito larvae. These water bodies were spread across 22 geo-referenced locations in Edo, Delta, and Anambra States (Table 1). These sites ranged from man-made (72.09%) to natural (27.91%) aquatic environments and comprised drains (48.84%), puddles (25.58%), abandoned car tyres (9.30%), buckets (4.65%), drums (4.65%), and stream margins (2.33%), as well as iron pan (2.33%) and concrete well (2.33%) (Figs. 1 and 2). A total of 1,778 larvae collected comprised 32.34% Anopheles and 67.66% culicine mosquitoes. Additional file 1 shows the number of water bodies (according to habitat type) that were positive for Anopheles larvae and those that were positive for culicine larvae. Meanwhile, Additional file 2 provides an account of water properties identified to predict Anopheles larvae in water body sites.
Anopheles species diversity
Sequencing and species identification
Overall, 528 out of the 575 field-collected Anopheles larvae were analyzed in PCR, with the success of DNA amplification for 382 larvae in S200 X6.1 gene-PCR and 42 larvae in ITS2-PCR; one larva was identified as coluzzii-gambiae s.s. hybrid in the S200 X6.1 gene-PCR. The remaining 104 samples that failed to amplify in S200 X6.1 gene-PCR and ITS2-PCR were analysed using molecular markers that target amplification of An. stephensi DNA; however, none of these 104 samples were amplified in endpoint-PCR except for the An. stephensi positive control was included in the reaction. Further, 78 samples were randomly selected from the 382 samples that were amplified in S200 X6.1 gene-PCR and all 42 samples that were amplified in ITS2-PCR were submitted to Sanger sequencing to confirm species identity. For S200 X6.1 sequences, NCBI BLAST search identified 58 sequences as An. coluzzii, 4 sequences as An. gambiae s.s., and 6 sequences as An. arabiensis, while 10 sequences had poor quality and were thus excluded from further analysis. For ITS2 sequences, 7 sequences having poor quality were discarded, while 3 sample sequences were identified in NCBI BLAST analysis as An. arabiensis and 32 sample sequences were identified as An. gambiae s.l.
Agreements between PCR-gel electrophoresis and amplicon sequencing for Anopheles species identification S200 X6.1 PCR and amplicon sequencing had near perfect agreement for the identification of An. coluzzii (Cohen’s Kappa K = 0.84) and An. gambiae s.s. (Cohen’s Kappa K = 0.90), but no agreement for the identification of An. arabiensis (Cohen’s Kappa K = 0.00). The lack of agreement between S200 X6.1 PCR and amplicon sequencing for An. arabiensis identification was due to the similarity of band sizes between An. gambiae ss (~ 249 bp) and An. arabiensis (~ 223 bp). The similarity resulted in An. arabiensis misidentification as An. gambiae s.s. in endpoint-PCR, but this misidentification was corrected in amplicon sequencing. Six (6) An. gambiae s.s. samples and 2 An. coluzzii samples so identified by PCR were shown by sequencing to be An. arabiensis and An. gambiae s.s., respectively. In an attempt to ensure that the study did not miss out on An. arabiensis, samples identified in endpoint PCR as An. gambiae s.s. were selected from different sites for amplicon sequencing.
Percentage identities and DNA sequence lengths Percentage identities of DNA sequences from the study when compared to GenBank DNA sequences ranged between 99.26% and 100% for the S200 X6.1 sequences with base-pair (bp) lengths of between 171bp and 180bp for An. arabiensis, 194bp and 210bp for An. gambiae s.s., and 407bp to 430bp for An. coluzzii. For ITS2 sequences, percentage identities ranged between 99.5% and 100% with base-pair lengths of between 508bp and 516bp for An. arabiensis and 399bp and 531bp for An. gambiae s.l.
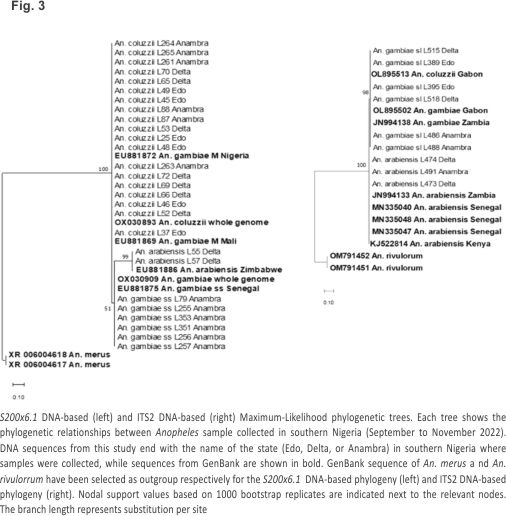
S200 X6.1 phylogeny Study DNA sequences of An. coluzzii on a maximum-likelihood phylogenetic tree (Fig. 3) clustered with GenBank DNA sequences of An. coluzzii whole genome (Accession No.: OX030893) and sequences of An. gambiae M molecular form from Mali (Accession No.: EU881869) and Nigeria (Accession No.: EU881872). Further, An. gambiae s.s. study sequences clustered with GenBank sequences of the whole genome of An. gambiae s.s. (Accession No.: OX030909) and an An. gambiae s.s. sequence from Senegal (Accession No.: EU881875). Lastly, DNA sequences of An. arabiensis from the study clustered with a sequence of the same species from Zimbabwe (Accession No.: Eu881886).
ITS2 phylogeny On the maximum-likelihood phylogenetic tree constructed from ITS2 sequences (Fig. 3), An. arabiensis study sequences clustered with An. arabiensis GenBank sequences from Senegal (Accession Nos. MN335047, MN335048, and MN335040), Kenya (Accession No.: KJ522814) and Zambia (Accession No.: JN994133), whereas study sequences of An. gambiae s.l. clustered with GenBank sequences of An. coluzzii from Gabon (Accession No.: OL895513) and An. gambiae from Gabon (Accession No. OL895502) and Zambia (Accession No. Jn994138).
Anopheles coluzzii, An. gambiae s.s., and An. arabiensis
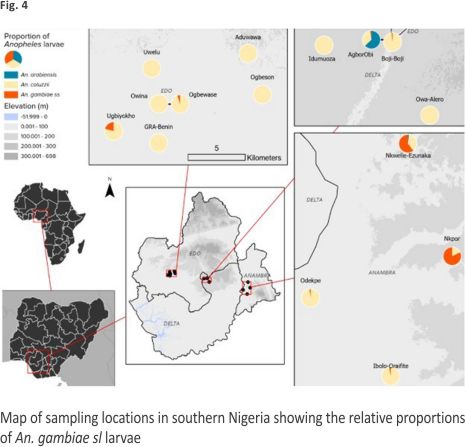
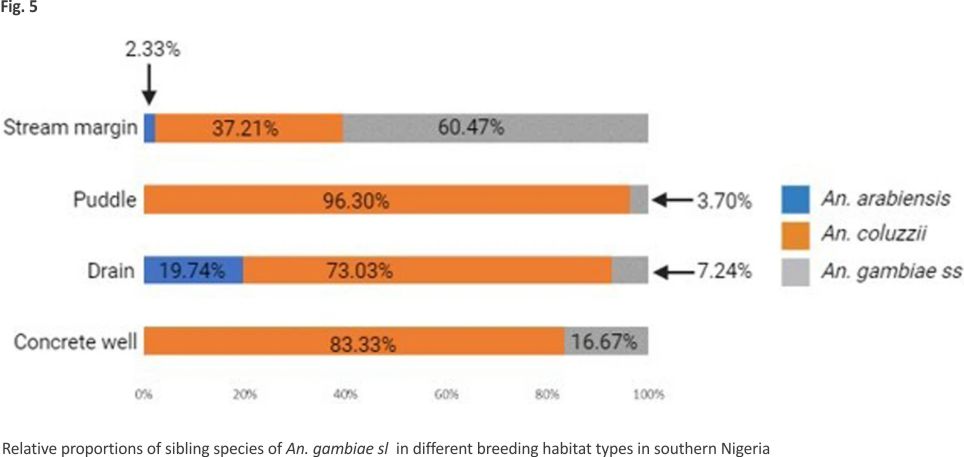
Overall distribution and proportions An. coluzzii occurred at more locations (68.18%, 15/22) compared to An. arabiensis (9.09%, 2/22) (Z-test: P = 0. 0002). However, the number of occurrence locations was similar between An. coluzzii and An. gambiae s.s. (36.36%, 8/22) (P = 0.07) and between An. arabiensis and An. gambiae s.s. (P = 0.07) (Fig. 4, Additional file 3). Overall, An. coluzzii larvae represented a greater proportion (80.51%, 314/ 390) in comparison to An. gambiae s.s. (11.54%, 45/390) (Z-test: P < 0.0001) and An. arabiensis (7.95%, 31/390) (P < 0.0001), whereas An. gambiae s.s. and An. arabiensis occurred at similar proportions (P = 0.1165). Anopheles coluzzii were detected mainly in puddles (57.96%, 182/314), and then in drains (35.35%, 111/314), stream margin (5.10%, 16/314), and a concrete well (1.596%, 5/314), while for An. gambiae s.s., detections were mainly in stream margin (57.78%, 26/45), followed by drains (24.44%, 11/45), puddles (15.56%, 7/45), and a concrete well (2.22%, 1/45) (Fig. 5). Anopheles arabiensis were detected at stream margin (3.23%, 1/31) and mainly in drains (96.77%, 30/31). Anopheles coluzzii and An. gambiae s.s. co-existed at 4 puddle sites and 3 drain sites, as well as in a concrete well. Meanwhile, An. coluzzii co-existed with An. arabiensis in drains at 2 sites, whereas all three species co-existed at the stream margin.
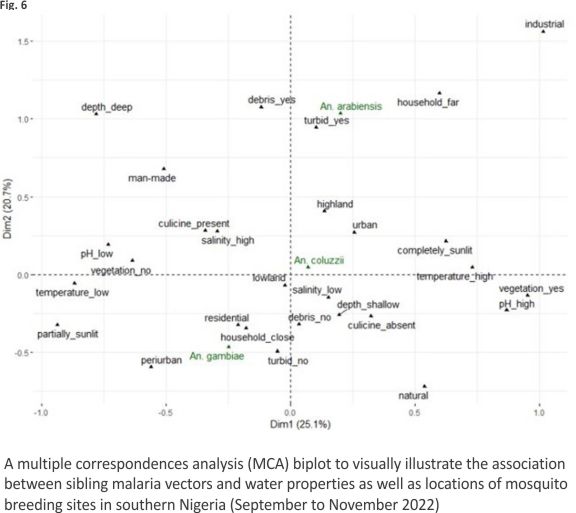
Multiple correspondence analysis The first two dimensions of the MCA explained 45.8% of variations in the properties of larval micro-habitats (Fig. 6). Twenty-five per cent and 20.7% of these variations were accounted for by dimensions 1 and 2 respectively. Sibling species of An. gambiae s.l. were separated more clearly along dimension 2 than dimension 1. Among water properties, water turbidity and the presence of debris in water made greater contributions to variations on dimension 2. The vector An. gambiae s.s. occupied the negative axis of dimension 2, whereas An. coluzzii and An. arabiensis occupied the positive axis of the same dimension (Fig. 6).
The separation of sibling species on the MCA biplot was more apparent for An. gambiae s.s. and An. arabiensis than for any other pair of the sibling vectors (Fig. 6). On dimension 2 of the biplot and more than the other vectors, An. gambiae s.s. showed close association with non-turbid water bodies and aquatic environments without debris and with partial exposure to sunlight, while also having closer association with human dwellings and periurban locations. Whereas the association of An. coluzzii with water properties on dimension 2 was less clear, An. arabiensis displayed close associations with turbid water, as well as water bodies containing debris and those far from households.
Random Forest classification
On variable importance plots (Additional file 4), the presence of culicine mosquitoes and debris, and habitat type and altitude were stronger predictors of Anopheles larvae and An. coluzzii presence (RF Accuracy 72.09%), whereas the presence of An. gambiae s.s. depended more on turbidity, exposure to sunlight, and the presence of culicine mosquitoes (RF Accuracy 83.72%) and An. arabiensis depended on culicine presence, temperature, and location (residential vs industrial) (RF Accuracy 93.02%).
Odds ratio analysis
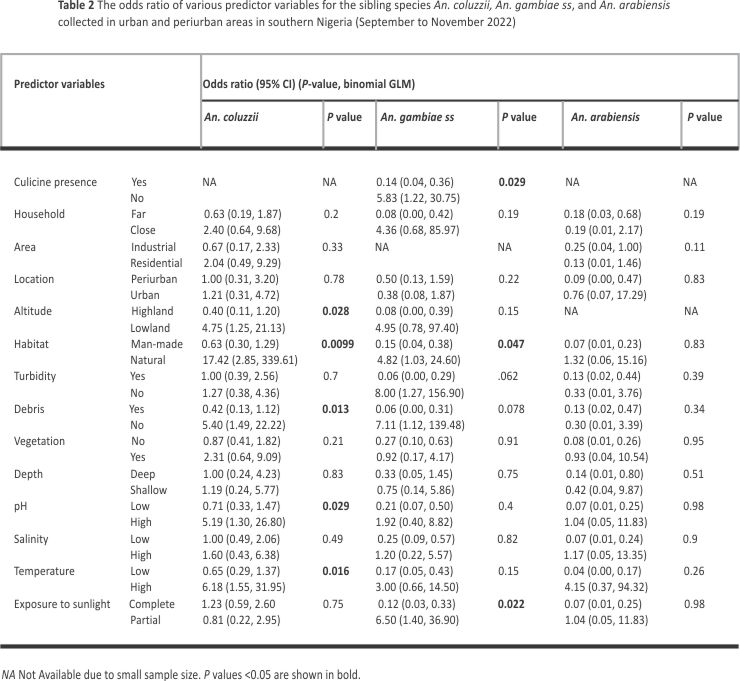
The odds of An. coluzzii detection was greater in natural habitats (OR: 17.42, 95% CI 2.85, 339.61) and water bodies without debris (OR: 5.40, 95% CI 1.49, 22.22). These odds were also greater in lowlands (OR: 4.74, 95% CI 1.25, 21.13) and aquatic environments with relatively high temperatures (OR: 6.18, 95% CI 1.55, 31.95) and pH (OR: 5.19, 95% CI 1.30, 26.80) (Table 2).
Odds of An. gambiae s.s. detection was greater in water bodies with partial rather than complete exposure to sunlight (OR: 6.50, 95% CI 1.40, 36.90), as well as in natural habitats (OR: 4.82, 95% CI 1.03, 24.60) and water bodies without culicine mosquitoes (OR: 5.83, 95% CI 1.22, 30.75) (Table 2).
Based on multivariate binomial regression modelling, topographic altitude and habitat exposure to sunlight respectively predicted the presence of An. coluzzii and An. gambiae s.s. in water bodies (Additional file 5). Habitat type also predicted the presence of An coluzzii and An. gambiae s.s. in the multivariate model analysis. The presence of An. arabiensis in water bodies was not assessed in multivariate regression due to the small sample size.
Mean proportions
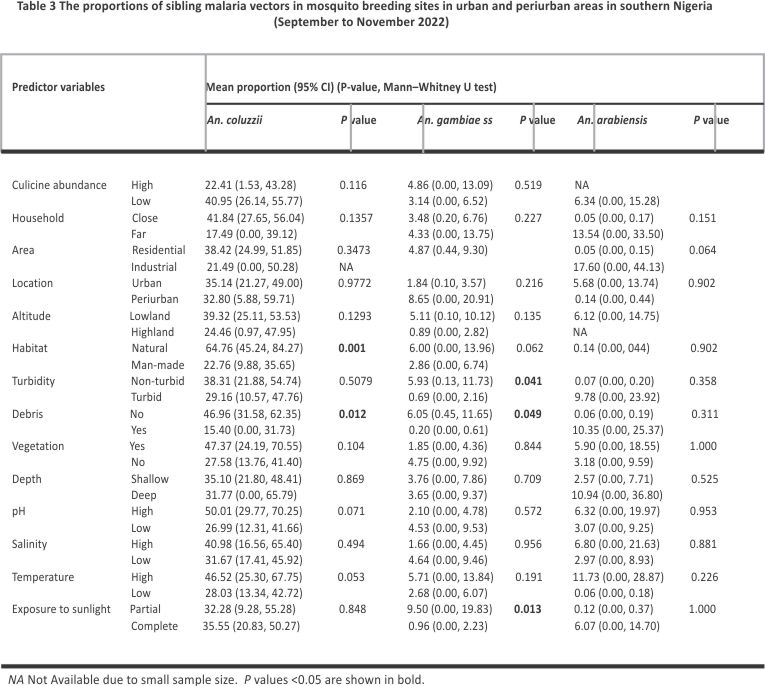
The relative mean proportion of An. coluzzii was greater in natural habitats (64.76, 95% CI 45.24, 84.27) and debris-free water bodies (46.96, 95% CI 31.58, 62.35) (Table 3). Anopheles gambiae s.s. also occurred in a greater proportion in debris-free water bodies (6.05, 95% CI 0.45, 11.65), as well as in water bodies that are non-turbid (5.93, 95% CI 0.13, 11.73) and partially sunlit (9.50, 95% CI 0.00, 19.83) (Table 3).
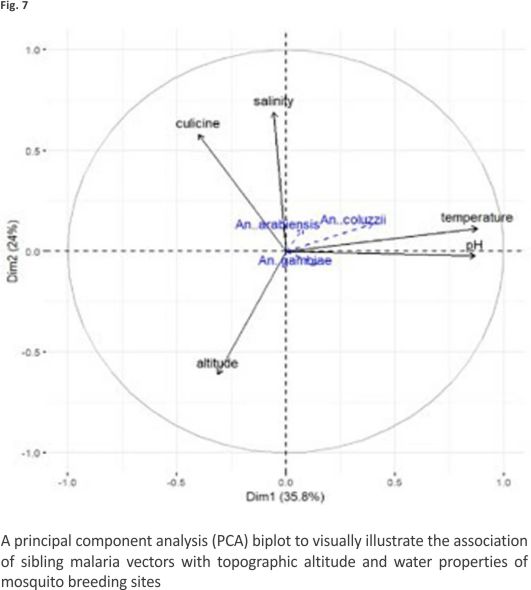
Proportions of An. coluzzii were negatively associated with altitude (ρ = − 0.33, P = 0.033) and positively associated with temperature (ρ = 0.36, P = 0.019). Figure 7 shows the direction and magnitude of these associations. Among predictor variables, only habitat exposure to sunlight could predict the proportions of An. gambiae s.s. in multivariate quasibinomial regression models (Additional file 5). No predictor variable was identified to statistically predict the proportions of An. coluzzii in multivariate models. The association between predictor variables and proportions of An. arabiensis could not be assessed in multivariate regression due to the small sample size.
Haplotype diversity of An. coluzzii populations based on DNA sequences of the S200 X6.1 gene
A total of 58 An. coluzzii DNA sequences (391 bp) were analysed to assess the possible effects of breeding habitat choices on genetic variations in An. coluzzii populations. The DNA sequences of An. coluzzii consisted of 6 haplotypes (H1 to H6), with a diversity (Hd) of 0.62 (Additional file 6). These haplotypes were segregated at 5 polymorphic sites, in addition to one nucleotide deletion in one of the study sequences (GenBank Accession Number: OR700036).
Additional file 7 shows the nucleotide positions of these segregations about the An. coluzzii Ngousso genome hosted in VectorBase 38. The nucleotide substitution A > G was the most common mutation while the nucleotide substitution A > C was the least common mutation. The H1 haplotype was more likely to occur in urban locations (OR: 3.80, 95% CI 1.21, 13.62) than in periurban locations (OR: 0.29, 95% CI 0.10, 0.74) (P = 0.028). Aside from this, no other association was detected between haplotype and geographical location or property of water bodies. Further analysis revealed low genetic variations between An. coluzzii populations that selected different breeding sites (Additional file 8).
Discussion
This study characterized the species diversity and breeding habitat choices of Anopheles malaria vectors in selected urban and periurban areas in southern Nigeria. Wild-caught Anopheles mosquito larvae comprised An. coluzzii, An. gambiae s.s., and An. arabiensis with preferred breeding sites in lowland, partially sunlit, and turbid water bodies, respectively. Furthermore, An. coluzzii and An. gambiae s.s. showed close association with breeding sites within 500 m of households, whereas An. arabiensis were associated with breeding sites outside 500 m of households.
Similar to findings in Ghana’s Cape Coast in West Africa 2, An. coluzzii occurred over a wider spatial range and at higher proportions compared to other sibling species. This may be explained by the adaptation of An. coluzzii larvae to predator pressure in the wild and the vector’s ability to outcompete sibling species in natural environments 39. Most mosquito breeding grounds in southern Nigeria had dried up during the dry season campaign. This left behind permanent breeding grounds that favour the proliferation of An. coluzzii and, as in previous work 7, contributed to the increased collection of this vector species.
The overexpression of detoxification genes by An. coluzzii has been demonstrated to enable larval individuals to exploit polluted urban breeding sites in Central Africa 40 and may further explain the high odds of An. coluzzii in urban and periurban water bodies in southern Nigeria. Findings revealed that An. coluzzii larvae were more likely to occur in water bodies in the lowlands than in the highlands. Except for a few cases where broad flat surfaces provided breeding places for mosquitoes in highlands 41, water bodies are more stable for mosquito breeding activities in lowlands. Also, warm temperatures in lowlands are favourable for mosquito larval survival and assist in accelerating rates of larval development 42,43,44.
Even though sampling did not include adult mosquitoes, it is likely that An. coluzzii dominated adult populations of malaria vectors during the predominantly dry sampling period in southern Nigeria and this contributed to increase collections of An. coluzzii larvae in study locations. Interestingly, earlier studies in similar ecologies that were conducted during dry periods of the year when temperatures were high alluded to the dominance of An. coluzzii among adult Anopheles mosquitoes 45, 46. Under high-temperature conditions, adult An. coluzzii are more able than other sibling vectors to minimize water loss and they do this by a variety of methods including altering the chemical compositions of cuticular hydrocarbons 47.
Anopheles gambiae s.s. have the behaviour of ovipositing in temporary, rain-dependent, small water collections such as puddles and hoof-prints 6. As the evaporation of small water collections is likely to occur over shorter periods during dry seasons, it seems that An. gambiae s.s. in southern Nigeria have developed a strategy that allows more time to complete the development of immature stages before breeding habitats completely dry up. This strategy, as the study results suggest, involves An. gambiae s.s. preference for water collections that are partially rather than completely exposed to sunlight. The present study therefore hypothesizes that direct and complete exposure to sunlight could hasten evaporation of temporary water bodies where An. gambiae s.s. breed and result in the death of immature mosquitoes before they reach adulthood.
For multiple reasons, wild-caught larvae in Delta and Anambra were not expected to include the outdoor-biting mosquito An. arabiensis. Firstly, An. arabiensis typically inhabit arid savannah landscapes and are often absent in field collections of adult or larval mosquitoes in the humid rainforest zone of southern Nigeria 48. Secondly, except on very few occasions 49,50,51, past and recent surveys in Delta and Anambra States have failed to detect An. arabiensis in field campaigns 14,17,52. Possible reasons for the absence of An. arabiensis in these earlier campaigns include but are not limited to the possibility that the vector was simply not present in sampling areas, or that investigators focused samplings on adult mosquitoes indoors and identified mosquitoes using less-sensitive techniques that are incapable of teasing apart sibling species.
In separate studies in Burkina Faso in West Africa 7 and Kenya in East Africa 53, An. arabiensis had its highest abundance during the dry season in October about the same time when An. gambiae s.s. had its lowest abundance. Mosquito larval collection during a similar period of the year likely increased the chances of An. arabiensis detection. This mosquito species is zoophilic and occurs close to livestock 54, thus it is not surprising that mosquito sampling led to the collection of many An. arabiensis larvae in water bodies in Agbor-Obi (Delta State) where there are several pockets of livestock-keeping areas.
The association between An. arabiensis and livestock production has been confirmed in a plethora of studies in sub-Saharan Africa 55,56,57 and is based on the fact that female mosquitoes, usually, restrict flight activities to places near animal blood meal hosts and oviposit in water bodies nearby. This could also explain why An. coluzzii and An. gambiae s.s., being anthropophilic mosquitoes, had almost all their breeding sites close to human residence within 500 m of households. In the Suba District in Kenya, households where > 90% of adult An. gambiae s.s. were collected and also had larval sites within 300 m 58. By breeding near blood meal hosts, female mosquitoes conserve flight energy and enable young adult progenies to easily access blood meals shortly after emergence.
Contrary to their choice of clean water bodies (see review by 48,57, An. arabiensis larvae occurred in polluted water in drainages in Agbor-Obi. Anopheles arabiensis larvae had also been found in polluted urban drainages and irrigation canals in the Khartoum State of Sudan 59. Furthermore, in Bobo-Dioulasso (Burkina Faso) where An. arabiensis has adapted to breeding in the polluted Houet River and can therefore transmit malaria throughout the dry season 60,61, the vector species increased in composition among malaria vectors from 3 to 90% 60 for two decades. Detections of An. arabiensis in these types of breeding places continues to increase 62,63, signalling a persistent and continuous adaptation of An. arabiensis larvae to polluted water bodies in urban areas, thereby promoting malaria transmission throughout the dry season.
In Central Africa, larvae of An. arabiensis that developed in organic wastewater developed faster, resulting in adults that had longer longevity and larger phenotypic sizes, as well as increased resistance to insecticides 64. Data on the association between An. arabiensis choice of breeding habitats and insecticide resistance traits are currently sparse in southern Nigeria. However, preliminary results from ongoing insecticide resistance studies in Agbor-Obi indicate the presence of the pyrethroid-resistant mutation L995F in larvae of An. arabiensis from polluted aquatic environments [unpublished data]. However, investigators are yet to find positive cases of L995F in An. arabiensis from clean water pools along stream margins in Nkwelle-Ezunaka (Anambra State).
In line with findings from the survey in southern Nigeria, a study in central Ethiopia identified An. arabiensis in turbid water collections 65. Similar observations of malaria vector preference for turbid water bodies were made in Tanzania 66. However, the species identity of mosquitoes was not determined. Anopheles mosquitoes typically avoid turbid for clean water for the reason that suspended insoluble particles interfere with larvae ingestion of food materials 65. These particles also limit sunlight penetration of water and consequently, slow down the production of aquatic microphyte food materials for mosquito larvae. These may have been responsible for the avoidance of turbid water bodies by An. gambiae s.s. and An. coluzzii in southern Nigeria.
However, turbidity may have less effect on An. arabiensis where larvae have adequate access to food materials. In the Ye-Ebiyo et al. 65 study in central Ethiopia, An. arabiensis were unaffected by turbidity of water bodies but only when larval sites were close to flowering maize plants providing pollen grains for larvae nourishment. In the present study, turbid water bodies that contained An. arabiensis larvae also contained organic debris. As Jeanrenaud et al. 64 observed in Cameroon, these organic wastes probably served as food for An. arabiensis larvae in southern Nigeria.
The Asian urban malaria vector An. stephensi invaded Africa in 2012 67 and has since been expanding its spatial range in the continent 68, with the most recent detections of the vector made in the West Africa sub-region 51,69, first in 2020 in Gombe in northern Nigeria 51. Due to its potential to drive malaria outbreaks 67,70, An. stephensi surveillance has received increased attention in areas of potential invasion. Molecular analysis in the present study screened Anopheles larval samples for An. stephensi because some samples could not be identified by molecular markers used in An. gambiae identification; similar experiences of molecular markers failing to identify wild samples of Anopheles mosquitoes led to the first reports of An. stephensi in different locations in Africa 71. Moreover, Sinka and colleagues 72 identified the study area in southern Nigeria among places in West Africa where ecological conditions are favourable for the invasion and establishment of An. stephensi.
Furthermore, given An. stephensi zoophagic habits 73, it is possible that the frequent pastoralists’ movement of livestock from Gombe and neighbouring locations in northern Nigeria to grazing fields and slaughterhouses in southern Nigeria could provide a route for and facilitate the southward spread of An. stephensi. In this study, mosquito larval samplings were carried out during the wet-dry season interface at a time when An. stephensi occur in high abundance 72 and with a majority of larval sampling sites comprising man-made water containers where the vector species prefers to breed in urban locations 11. Considering that the larval sampling strategy maximized opportunities for An. stephensi detection, the non-report of the vector among the study mosquito samples therefore suggests its absence in the sampling area and likely slow spread in the country Nigeria. However, An. stephensi possesses the potential for rapid spatial distribution. This has been demonstrated in East Africa, where the vector species was detected in five countries within 10 years 68 and in West Africa, where it was recently detected in Accra Ghana 69, just less than 3 years after the initial detection in northern Nigeria 51.
The An. coluzzii population in southern Nigeria was moderately genetically diverse, with a haplotype diversity index of 0.62. This suggests that vector control interventions are currently not optimally effective at reducing An. coluzzii abundance in the study area; otherwise, vector populations would have presented with low genetic diversity. High genetic diversity of An. coluzzii was attributed to variations in the ecology of larval development sites along the Gambian River in West Africa 74. In southern Nigeria, An. coluzzii that developed in periurban larval sites had slightly more haplotypes than those in urban sites. However, and possibly as an indication of An. coluzzii’s attempt to adapt to otherwise less favourable conditions in urban ecological landscapes, one of the two dominant haplotypes detected in An. coluzzii occurred in close association with the urban vector population. Due to the small sample size, the use of a less informative molecular marker, and the restriction of nucleotide sequencing to limited regions of the genome, it was difficult to adequately assess genetic divergence between An. coluzzii populations in the present study.
Turbidity metres or Secchi disks are recommended for reliable assessment of water turbidity; hence investigators admit that the method of assessing water turbidity based on physical observations may have been less accurate. Physical observation to assess water turbidity could also be subjective; however, in the present study, water turbidity assessment by the same person helped to address this challenge in the field. Further, the study did not systematically evaluate the effects of biological factors. Some of these biological factors, for example, the presence of predators, have been shown in previous studies to affect Anopheles larvae in aquatic environments (reviewed in 6,75. However, the relative proportions of Anopheles larvae and the fact that sibling vectors rarely co-existed in water body sites suggest that sibling mosquitoes could be engaging in some sort of inter-specific competition for resources 39,76). The stream margin in Nkwelle-Ezunaka (Anambra) was the only place where An. arabiensis, An. gambiae s.s. and An. coluzzii occurred together in a single water body. It could be that mosquito larvae because they are less crowded in large breeding sites such as stream margins, are less likely to engage in resource competition. Still, on mosquito interactions, Anopheles and culicine larvae had inverse associations in southern Nigeria. The inverse association observed between Anopheles and culicine larvae corroborates the principle that gravid dipterans typically avoid breeding places already exploited by conspecific and heterospecific females 77. This behaviour has been reported in An. gambiae s.s. 78 and aims to ensure adequate food resources for potential immature progenies and thus enhance the biological fitness of adult progenies.12
In conclusion, the study reports different breeding habitat choices for three sibling malaria vectors in southern Nigeria. The dominant vector An. coluzzii prefer breeding sites in lowlands while An. gambiae s.s. prefer sites that are partially rather than completely exposed to sunlight. In contrast to An. arabiensis that display association with man-made sites outside 500 m of households, An. coluzzii and An. gambiae s.s. have a high likelihood of breeding in natural sites within 500 m of households. These findings suggest that An. coluzzii and An. gambiae s.s. are more likely than An. arabiensis to infect humans in residential places where the vectors co-exist 79. And as they are typically indoor feeders, An. coluzzii and An. gambiae s.s. have a greater chance of contacting and being killed by treated bed nets.
There are ongoing efforts by State governments to upscale the distribution and encourage the use of pyrethroid-treated bed nets for malaria vector control in southern Nigeria 19. However, An. arabiensis, being an outdoor feeder and capable of deriving bloodmeals from multiple vertebrates in addition to humans, has a lower opportunity of encountering treated bed nets. In East Africa, treated bed nets helped control An. gambiae s.s., but An. arabiensis were only slightly affected, thus leaving behind a post-intervention phase of residual malaria transmission by An. arabiensis 80. Findings of pyrethroid resistance mutations in An. arabiensis in the study area [unpublished data] and escalations of insecticide resistance in An. coluzzii in southern Nigeria 18 further dampens the prospect of vector control using treated bed nets.
Malaria control programmes in southern Nigeria could leverage findings from the present study in designing targeted larval control interventions. Across sub-Saharan Africa, larval control interventions have been explored to reduce the population abundance of An. arabiensis and a couple of other mosquito species that transmit infections, irrespective of vector biting location (indoors or outdoors) or insecticide resistance status (resistant or susceptible) 81,82. Larval control is particularly useful in the context of southern Nigeria where bed net interventions are having limited effects on malaria vectors. Larval control interventions to reduce human malaria transmission are easier to implement during the dry season when several water collections providing breeding places for Anopheles vectors have dried up 12. As rainfall amounts decrease, mosquito breeding activities are concentrated to fewer water collections, which, if targeted in larval control interventions, could improve the goal of reducing malaria risks in periods of little or no rainfall. The present study has identified potential sites for larval control interventions during such periods in southern Nigeria. It has also reported the absence of An. stephensi in selected urban and periurban locations in the area. However, southern Nigeria is exposed to An. stephensi invasion is a travel destination for land, air, and sea transport from places where the vector species has already established its presence. This raises a need for the National Malaria Control Programme and relevant health authorities at the subnational levels to create a system for the surveillance of urban and periurban locations for An. stephensi. Such surveillance should focus on man-made mosquito breeding sites where larval interventions could help to slow the spread and proliferation of An. stephensi in the event of an invasion.
Availability of data and materials
All data generated and/or analysed during this study are included in this published article and the supplementary files. The raw dataset will be made available upon reasonable request to the corresponding author. DNA sequences generated from the study have been deposited in the GenBank database using the Accession Numbers OR700033 to OR700102, and OR717035 to OR717056.
Abbreviations
BP: Base-pair
CI: Confidence interval
DNA: Deoxyribonucleic Acid
GLM: Generalized Linear Model
Hd: Haplotype diversity
ITS2: Internal Transcribed Spacer-2
MCA: Multiple Correspondence Analysis
MDG: Mean Decrease Gini
OR: Odds ratio
PCA: Principal Components Analysis
PCR: Polymerase chain reaction
RF: Random Forest
SINE: Short Interspersed Elements
℃: Degree Celsius
WHO: World Health Organization
RDT: Rapid Diagnostic Test
mm: Millimeter
MJ: Median-Joining
References
1. WHO. World Malaria Report 2022. Geneva: World Health Organization; 2022.
2. Kudom AA, Mensah BA, Agyemang TK. Characterization of mosquito larval habitats and assessment of insecticide-resistance status of Anopheles gambiae sensu lato in urban areas in southwestern Ghana. J Vector Ecol. 2012;37:77–82.
3. Emidi B, Kisinza WN, Mmbando BP, Malima R, Mosha FW. Effect of physicochemical parameters on Anopheles and Culex mosquito larvae abundance in different breeding sites in a rural setting of Muheza Tanzania. Parasit Vectors. 2017;10: 304.
4. Tennessen JA, Ingham VA, Toé KH, Guelbéogo WM, Sagnon NF, Kuzma R, et al. A population genomic unveiling of a new cryptic mosquito taxon within the malaria-transmitting Anopheles gambiae complex. Mol Ecol. 2021;30:775 –90.
5. Irish S, Kyalo D, Snow R, Coetzee M. Anopheles species present in countries in sub-Saharan Africa and associated islands. Zootaxa. 2019; 4747:401–49.
6. Lehmann T, Diabate A. The molecular forms of Anopheles gambiae: a phenotypic perspective. Infect Genet Evol. 2008;8:737–46.
7. Gimonneau G, Pombi M, Choisy M, Morand S, Dabiré RK, Simard F. Larval habitat segregation between the molecular forms of the mosquito Anopheles gambiae in a rice field area of Burkina Faso. West Africa Med Vet Entomol. 2012;26:9–17.
8. Touré YT, Petrarca V, Traore SF, Coulibaly A, Maiga HM, Sankare O, et al. The distribution and inversion polymorphism of chromosomally recognized taxa of the Anopheles gambiae complex in Mali. West Africa Parassitologia. 1998; 40: 477–511.
9. Kamdem C, Tene Fossog B, Simard F, Etouna J, Ndo C, Kengne P, et al. Anthropogenic habitat disturbance and ecological divergence between incipient species of the malaria mosquito Anopheles gambiae. PLoS ONE. 2012;7: e39453.
10. Akpodiete NO, Tripet F. Laboratory and microcosm experiments reveal contrasted adaptive responses to ammonia and water mineralization in aquatic stages of the sibling species Anopheles gambiae (sensustricto) and Anopheles coluzzii. Parasit Vectors. 2021;14:17.
11. Hemming-Schroeder E, Ahmed A. Anopheles stephensi in Africa: vector control opportunities for cobreeding An. stephensi and Aedes arbovirus vectors. Trends Parasitol. 2023;39: 86–90.
12. WHO. Larval source management: a supplementary malaria vector control measure: an operational manual. Geneva: World Health Organization; 2013.
13. Okorie PN, McKenzie FE, Ademowo OG, Bockarie M, Kelly-Hope L. Nigeria Anopheles vector database: an overview of 100 years’ research. PLoS ONE. 2011;6: e28347.
14. The PMI Vector Link Project. The PMI VectorLink Nigeria Project Annual Entomology Report, October 2020– September 2021. Rockville: VectorLink, Abt Associates Inc.; 2022.
15. Takken W. Do insecticide-treated bednets have an effect on malaria vectors? Trop Med Int Health. 2002;7:1022–30.
16. Russell TL, Govella NJ, Azizi S, Drakeley CJ, Kachur SP, Killeen GF. Increased proportions of outdoor feeding among residual malaria vector populations following increased use of insecticide-treated nets in rural Tanzania. Malar J. 2011;10:80.
17. Awolola TS, Adeogun A, Olakiigbe AK, Oyeniyi T, Olukosi YA, Okoh H, et al. Pyrethroids resistance intensity and resistance mechanisms in Anopheles gambiae from malaria vector surveillance sites in Nigeria. PLoS ONE. 2018;13: e0205230.
19. Muhammad A, Ibrahim SS, Mukhtar MM, Irving H, Abajue MC, Edith NM, et al. High pyrethroid/DDT resistance in major malaria vector Anopheles coluzzii from Niger Delta of Nigeria is probably driven by metabolic resistance mechanisms. PLoS ONE. 2021;16: e0247944.
20. WHO African Region. Report on malaria in Nigeria. Geneva: WHO; 2022. p. 1–95.
21. Climate Change Knowledge Portal. https://climateknowledgeportal.worldbank.org/country/nigeria/climate-data-historical#:~:text =Annual% 20rainfall%20can%20 reach%20 up%20to%20about% 201200,rest%20of%20the%20year%20is%20hot%20and%20dry.
22. Musapa M, Kumwenda T, Mkulama M, Chishimba S, Norris DE, Thuma PE, Mharakurwa S. A simple Chelex protocol for DNA extraction from Anopheles spp. J Vis Exp. 2013;9: e3281.
23. Santolamazza F, Mancini E, Simard F, Qi Y, Tu Z, della Torre A. Insertion polymorphisms of SINE200 retrotransposons within speciation islands of Anopheles gambiae molecular forms. Malar J. 2008;7: 163 .
24. Beebe NW, Saul A. Discrimination of all members of the Anopheles punctulatus complex by polymerase chain reaction-restriction fragment length polymorphism analysis. Am J Trop Med Hyg. 1995; 53:478–81.
25. Singh OP, Kaur T, Sharma G, Kona MP, Mishra S, Kapoor N, Mallick PK. Molecular tools for early detection of invasive malaria vector Anopheles stephensi mosquitoes. Emerg Infect Dis. 2023;29:36.
26. Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In: Nucleic acids symposium series 1999;41: 95–98.
27. The National Center for Biotechnology Information NCBI. https://blast. ncbi.nlm.nih.gov/Blast.cgi.
28. Clustal Omega Multiple Sequence Alignment. https://www.ebi.ac. uk/Tools/msa/clustalo/.
29. Lefort V, Longueville JE, Gascuel O. SMS: smart model selection in PhyML. Mol Biol Evol. 2017;34:24 22– 4.
30. Hasegawa M, Kishino H, Yano TA. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 1985;22:160–74.
31. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547.
32. Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, et al. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol. 2017;34:3299–302.
33. Bandelt HJ, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48.
34. Leigh JW, Bryant D. POPART: full-feature software for haplotype network construction. Methods Ecol Evol. 2015;6:1110–6.
35. Breiman L, Cutler RA. Random forests machine learning. J Clin Microbiol. 2001;2:199–228.
36. Kassambara A. Practical guide to principal component methods in R: PCA, M (CA), FAMD, MFA, HCPC, factoextra. Sthda. 2017.
37. Lenth R, Lenth MR. Package ‘lsmeans.’ Am Stat. 2018;34:216–21.
38. R Core Team. R: A language and environment for statistical computing. Vienna: R Core Team; 2010.
39. VectorBase Bioinformatics Resources for Invertebrate Vectors of Human Pathogens. https://vectorbase.org/ vectorbase/app).
40. Diabaté A, Dabiré RK, Heidenberger K, Crawford J, Lamp WO, Culler LE, Lehmann T. Evidence for divergent selection between the molecular forms of Anopheles gambiae: role of predation. BMC Evol Biol. 2008; 8:5.
41. Cassone BJ, Kamdem C, Cheng C, Tan JC, Hahn MW, Costantini C, Besansky NJ. Gene expression divergence between malaria vector sibling species Anopheles gambiae and An. coluzzii from rural and urban Yaoundé Cameroon. Mol Ecol. 2014;23: 2242–59.
42. Atieli HE, Zhou G, Lee MC, Kweka EJ, Afrane Y, Mwanzo I, et al. Topography as a modifier of breeding habitats and concurrent vulnerability to malaria risk in the western Kenya highlands. Parasit Vectors. 2011;4: 241.
43. Tuno N, Okeka W, Minakawa N, Takagi M, Yan G. Survivorship of Anopheles gambiae sensu stricto (Diptera: Culicidae) larvae in western Kenya highland forest. J Med Entomol.2005; 42:270–7.
44. Tene Fossog B, Antonio-Nkondjio C, Kengne P, Njiokou F, Besansky NJ, Costantini C. Physiological correlates of ecological divergence along an urbanization gradient: differential tolerance to ammonia among molecular forms of the malaria mosquito Anopheles gambiae. BMC Ecol. 2013;13:1.
45. Tene Fossog B, Ayala D, Acevedo P, Kengne P, Ngomo Abeso Mebuy I, et al. Habitat segregation and ecological character displacement in cryptic African malaria mosquitoes. Evol Appl. 2015;8:326–45.
46. Fahmy NT, Villinski JT, Bolay F, Stoops CA, Tageldin RA, Fakoli L, et al. The seasonality and ecology of the Anopheles gambiae complex (Dipetra: Culicidae) in Liberia using molecular identification. J Med Entomol. 2015; 52:475–82.
47. Akogbéto MC, Salako AS, Dagnon F, Aïkpon R, Kouletio M, Sovi A, et al. Blood feeding behaviour comparison and contribution of Anopheles coluzzii and Anopheles gambiae, two sibling species living in sympatry, to malaria transmission in Alibori and Donga region, northern Benin. West Africa Malar J. 2018; 17:307.
48. Arcaz AC, Huestis DL, Dao A, Yaro AS, Diallo M, Andersen J, et al. Desiccation tolerance in Anopheles coluzzii: the effects of spiracle size and cuticular hydrocarbons. J Exp Biol. 2016;219: 1675–88.
49. Sinka ME, Bangs MJ, Manguin S, Coetzee M, Mbogo CM, Hemingway J, et al. The dominant Anopheles vectors of human malaria in Africa, Europe and the Middle East: occurrence data, distribution maps and bionomic précis. Parasit Vectors. 2010;3:117.
50. Onyabe DY, Conn JE. The distribution of two major malaria vectors, Anopheles gambiae and Anopheles arabiensis, in Nigeria. Mem Inst Oswaldo Cruz. 2001;96:1081–4.
51. Okwa OO, Akinmolayan FI, Carter V, Hurd H. Transmission dynamics of malaria in Nigeria. Ann Afr Med. 2009; 8:1–9.
52. Adeogun A, Babalola AS, Okoko OO, Oyeniyi T, Omotayo A, Izekor RT, et al. Spatial distribution and ecological niche modelling of geographical spread of Anopheles gambiae complex in Nigeria using real-time data. Sci Rep. 2023;13: 13679.
53. Chukwuekezie O, Nwosu E, Nwangwu U, Dogunro F, Onwude C, Agashi N, et al. Resistance status of Anopheles gambiae (s.l.) to four commonly used insecticides for malaria vector control in South-East Nigeria. Parasit Vectors. 2020;13:152.
54. Kweka EJ, Zhou G, Munga S, Lee MC, Atieli HE, Nyindo M, et al. Anopheline larval habitats seasonality and species distribution: a prerequisite for effective targeted larval habitats control programmes. PLoS ONE. 2012; 7: e52084.
55. Asale A, Duchateau L, Devleesschauwer B, Huisman G, Yewhalaw D. Zooprophylaxis as a control strategy for malaria caused by the vector Anopheles arabiensis (Diptera: Culicidae): a systematic review. Infect Dis Poverty. 2017;6: 160.
56. Charlwood JD, Edoh D. Polymerase chain reaction used to describe larval habitat use by Anopheles gambiae complex (Diptera: Culicidae) in the environs of Ifakara Tanzania. J Med Entomol. 1996;33:202–4.
57. Mahande A, Mosha F, Mahande J, Kweka E. Feeding and resting behaviour of malaria vector, Anopheles arabiensis with reference to zooprophylaxis. Malar J. 2007; 6:100.
58. Gone T, Balkew M, Gebre-Michael T. Comparative entomological study on ecology and behaviour of Anopheles mosquitoes in highland and lowland localities of Derashe District, southern Ethiopia. Parasit Vectors. 2014;7:483.
59. Minakawa N, Seda P, Yan G. Influence of host and larval habitat distribution on the abundance of African malaria vectors in western Kenya. Am J Trop Med Hyg. 2002; 67:32–8.
60. Azrag RS, Mohammed BH. Anopheles arabiensis in Sudan: a noticeable tolerance to urban polluted larval habitats associated with resistance to Temephos. Malar J. 2018;17:204.
61. Dabiré RK, Namountougou M, Sawadogo SP, Yaro LB, Toé HK, Ouari A, et al. Population dynamics of Anopheles gambiae sl in Bobo-Dioulasso city: bionomics, infection rate and susceptibility to insecticides. Parasit Vectors. 2012;5:127.
62. Soma DD, Kassié D, Sanou S, Karama FB, Ouari A, Mamai W, et al. Uneven malaria transmission in geographically distinct districts of Bobo-Dioulasso. Burkina Faso Parasit Vectors. 2018; 11:296.
63. Gordicho V, Vicente JL, Sousa CA, Caputo B, Pombi M, Dinis J, et al. First report of an exophilic Anopheles arabiensis population in Bissau City, Guinea-Bissau: recent introduction or sampling bias? Malar J. 2014;13: 423.
64. Fournet F, Adja AM, Adou KA, Dahoui M, Coulibaly B, Assouho KF, et al. First detection of the malaria vector Anopheles arabiensis in Côte d’Ivoire: urbanization in question. Malar J. 2022;21:275.
65. Jeanrenaud AC, Brooke BD, Oliver SV. Second generation effects of larval metal pollutant exposure on reproduction, longevity and insecticide tolerance in the major malaria vector Anopheles arabiensis (Diptera: Culicidae). Parasit Vectors. 2020;13:4.
66. Ye-Ebiyo Y, Pollack RJ, Kiszewski A, Spielman A. Enhancement of development of larval Anopheles arabiensis by proximity to flowering maize (Zea mays) in turbid water and when crowded. Am J Trop Med Hyg. 2003;68:748–52.
67. Sattler MA, Mtasiwa D, Kiama M, Premji Z, Tanner M, Killeen GF, et al. Habitat characterization and spatial distribution of Anopheles sp. Mosquito larvae in Dar es Salaam (Tanzania) during an extended dry period. Malar J. 2005;4:4.
68. Faulde MK, Rueda LM, Khaireh BA. First record of the Asian malaria vector Anopheles stephensi and its possible role in the resurgence of malaria in Djibouti. Horn of Africa Acta Trop. 2014;139:39–43.
69. WHO initiative to stop the spread of Anopheles stephensi in Africa. 2023 Update. 2023. https://www. who. int/publications/I/item/ WHO-UCN-GMP-2022.06. A
70. Afrane YA, Abdulai A, Mohammed AR, Akuamoah-Boateng Y, Owusu-Asenso CM, Sraku IK, et al. First detection of Anopheles stephensi in Ghana using molecular surveillance. Biorxiv. 2023. https: //doi.org/10.1101/2023.12.01. 569589.
71. Emiru T, Getachew D, Murphy M, Sedda L, Ejigu LA, Bulto MG, et al. Evidence for the role of Anopheles stephensi in the spread of drug and diagnosis-resistant malaria in Africa. Nat Med. 2023;29: 3203–11.
72. Ahmed A, Pignatelli P, Elaagip A, Hamid MM, Alrahman OF, Weetman D. Invasive malaria vector Anopheles stephensi mosquitoes in Sudan, 2016–2018. Emerg Infect Dis. 2021;27:2952.
73. Sinka ME, Pironon S, Massey NC, Longbottom J, Hemingway J, Moyes CL, et al. A new malaria vector in Africa: predicting the expansion range of Anopheles stephensi and identifying the urban populations at risk. Proc Natl Acad Sci USA. 2020; 117:24900–8.
74. Altahir O, AbdElbagi H, Abubakr M, Siddig EE, Ahmed A, Mohamed NS. Blood meal profile and positivity rate with malaria parasites among different malaria vectors in Sudan. Malar J. 2022; 21:124.
75. Caputo B, Nwakanma D, Caputo FP, Jawara M, Oriero EC, Hamid-Adiamoh M, et al. Prominent intraspecific genetic divergence within Anopheles gambiae sibling species triggered by habitat discontinuities across a riverine landscape. Mol Ecol. 2014;23:457 4 –89.
76. Kweka EJ, Zhou G, Gilbreath TM, Afrane Y, Nyindo M, Githeko AK, Yan G. Predation efficiency of Anopheles gambiae larvae by aquatic predators in western Kenya highlands. Parasit Vectors. 2011;4:128.
77. Schneider P, Takken W, McCall PJ. Interspecific competition between sibling species larvae of Anopheles arabiensis and An. gambiae. Med Vet Entomol. 2000;14:165–70.
78. Baleba SB, Torto B, Masiga D, Getahun MN, Weldon CW. Stable flies, Stomoxys calcitrans L. (Diptera: Muscidae), improve offspring fitness by avoiding oviposition substrates with competitors or parasites. Front Ecol Evol. 2020;8: 5.
79. Munga S, Minakawa N, Zhou G, Barrack OO, Githeko AK, Yan G. Effects of larval competitors and predators on oviposition site selection of Anopheles gambiae sensu stricto. J Med Entomol. 2014;43:221–4.
80. Midega JT, Smith DL, Olotu A, Mwangangi JM, Nzovu JG, Wambua J, et al. Wind direction and proximity to larval sites determines malaria risk in Kilifi District in Kenya. Nat Commun. 2012;3:674.
81. Bayoh MN, Mathias DK, Odiere MR, Mutuku FM, Kamau L, Gimnig JE, et al. Anopheles gambiae: historical population decline associated with regional distribution of insecticide-treated bed nets in western Nyanza Province. Kenya Malar J. 2010;9:6 2.
82. Killeen GF, Fillinger U, Knols BG. Advantages of larval control for African malaria vectors: low mobility and behavioural responsiveness of immature mosquito stages allow high effective coverage. Malar J. 2002;1:8.
83. Walker K, Lynch M. Contributions of Anopheles larval control to malaria suppression in tropical Africa: review of achievements and potential. Med Vet Entomol. 2007;21:2–21.
Acknowledgements
The authors are grateful to Sunday Lee Akpama and Philip Ogbeide Aihebholoria for assistance during the field collection of samples. The GIS Unit at Colorado State University USA helped to design the map showing the proportions of Anopheles larvae in sampling locations.
Funding
The work was funded from the start-up grant awarded to EHS by Colorado State University. Colorado State University had no role in the conceptualization, design, data collection, analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Centre for Vector-Borne Infectious Diseases, Department of Microbiology, Immunology, and Pathology, Colorado State University, Fort Collins, CO, USA
Faith I. Ebhodaghe, Irma Sanchez-Vargas, Brian D. Foy & Elizabeth Hemming-Schroeder
Department of Zoology, Faculty of Life Sciences, Ambrose Alli University, Ekpoma, Edo State, Nigeria
Clement Isaac
Contributions
FIE conceived the research. FIE, EHS, BDF, and CI designed the research. EHS supervised the study. FIE collected field samples. FIE and ISV performed lab work. FIE analysed the data. FIE drafted the manuscript. FIE, EHS, ISV, and BDF edited the manuscript. All authors read and approved the final version of the manuscript.
Corresponding author
Correspondence to Elizabeth Hemming-Schroeder.
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
All authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral concerning jurisdictional claims in published maps and institutional affiliations.
CREDITS: Ebhodaghe, F.I., Sanchez-Vargas, I., Isaac, C. et al. Sibling species of the major malaria vector Anopheles gambiae display divergent preferences for aquatic breeding sites in southern Nigeria. Malar J 23, 60 (2024). https://doi.org/10. 1186 /s12936-024-04871-9