Suttipat Srisutham, Kanokon Suwannasin, Rungniran Sugaram, Arjen M. Dondorp & Mallika Imwong
Abstract
Background
Copy number variations (CNVs) of the Plasmodium falciparum multidrug resistance 1 (pfmdr1), P. falciparum plasmepsin2 (pfplasmepsin2) and P. falciparum GTP cyclohydrolase 1 (pfgch1) genes are associated with anti-malarial drug resistance in P. falciparum malaria. Droplet digital PCR (ddPCR) assays have been developed for the accurate assessment of CNVs in several human genes. The aim of the present study was to develop and validate ddPCR assays for the detection of the CNVs of P. falciparum genes associated with resistance to anti-malarial drugs.
Methods
A multiplex ddPCR assay was developed to detect the CNVs in the pfmdr1 and pfplasmepsin2 genes, while a duplex ddPCR assay was developed to detect CNV in the pfgch1 gene. The gene copy number (GCN) quantification limit, as well as the accuracy and precision of the ddPCR assays, were determined and compared to conventional quantitative PCR (qPCR). In order to reduce the cost of testing, a multiplex ddPCR assay of two target genes, pfmdr1 and pfplasmepsin2, was validated. In addition, the CNVs of genes of field samples collected from Thailand from 2015 to 2019 (n = 84) were assessed by ddPCR and results were compared to qPCR as the reference assay.
Results
There were no significant differences between the GCN results obtained from uniplex and multiplex ddPCR assays for detection of CNVs in the pfmdr1 and pfplasmepsin2 genes (p = 0.363 and 0.330, respectively). Based on the obtained gene copy number quantification limit, the accuracy and per cent relative standard deviation (%RSD) value of the multiplex ddPCR assay were 95% and 5%, respectively, for detection of the CNV of the pfmdr1 gene, and 91% and 5% for detection of the CNV of the pfplasmepsin2 gene. There was no significant difference in gene copy numbers assessed by uniplex or duplex ddPCR assays regarding CNV in the pfgch1 gene (p = 0.276). The accuracy and %RSD value of the duplex ddPCR assay were 95% and 4%, respectively, regarding pfgch1 GCN. In the P. falciparum field samples, pfmdr1 and pfplasmepsin2 GCNs were amplified in 15% and 27% of samples from Ubon Ratchathani, Thailand, while pfgch1 GCN was amplified in 50% of samples from Yala, Thailand. There was 100% agreement between the GCN results obtained from the ddPCR and qPCR assays (κ = 1.00). The results suggested that multiplex ddPCR assay is the optional assay for the accurate detection of gene copy number without requiring calibration standards, while the cost and required time are reduced. Based on the results of this study, criteria for GCN detection by ddPCR analysis were generated.
Conclusions
The developed ddPCR assays are simple, accurate, precise and cost-effective tools for detection of the CNVs in the pfmdr1, pfplasmepsin2 and pfgch1 genes of P. falciparum. The ddPCR assay is a useful additional tool for the surveillance of anti-malarial drug resistance.
Background
Artemisinin-based combination therapy (ACT) is recommended as a front-line treatment for Plasmodium falciparum malaria, which remains an important infectious disease in tropical regions. However, the emergence and spread of resistance to artemisinin-based combinations and related drugs have resulted in poor curative rates, especially in Southeast Asia 1,2,3,4,5. Molecular surveillance is needed not only for the detection of mutations to the P. falciparum kelch gene, which are associated with artemisinin resistance3, but also molecular markers associated with the efficacy of other anti-malarial drugs. An increase in the P. falciparum multidrug resistance 1 (pfmdr1) GCN is associated with mefloquine resistance6, while an increase in the P. falciparum plasmepsin2 (pfplasmepsin2) GCN is associated with piperaquine resistance7,8. Moreover, amplification of the P. falciparum GTP cyclohydrolase 1 (pfgch1) GCN is linked to upregulation of the P. falciparum dihydrofolate reductase (pfdhfr) and P. falciparum dihydropteroate synthase (pfdhps) genes, which are associated with sulfadoxine-pyrimethamine resistance in Southeast Asia 9,10.
Quantitative PCR (qPCR) assays are conventionally used to assess the copy number variations (CNVs) of genes related to drug resistance in P. falciparum malaria (i.e. pfmdr1 6, pfplasmepsin2 7, and pfgch1 11). Alternatively, droplet digital PCR (ddPCR) technology was developed to measure CNVs and to provide highly precise measurements of the concentrations of target and reference genes in DNA samples 12,13, as well as tolerance to PCR inhibitors, such as heparin 14, and to generate calibration curves to determine the GCNs of target sequences 15,16. The ddPCR assay is based on water-oil emulsion droplet technology used for detection and quantification of target gene 17. The ddPCR reaction contains the ddPCR reagent, DNA samples, primers and fluorescent probe. All components are divided into around 15,000–20,000 droplets using the droplet generator. Each droplet may contain one, more than one or no copies of the DNA target 12,13,18. After 40 cycles of the standard PCR reaction, DNA targets in each droplet are amplified and then analysed by a droplet reader. The DNA target concentration is calculated from the number of positive and negative droplets using Poisson statistics 12. The ddPCR assay has been developed for accurate detection of CNVs in human genes associated with various human genetic diseases 19,20,21. In addition, a ddPCR assay was developed and validated for the detection and quantification of Plasmodium species based on the 18S rRNA gene sequence 22,23, but this method has not yet been validated for the detection of the CNVs of genes associated with resistance to anti-malarial drugs.
In the present study, ddPCR assays were developed and validated for quantification of the CNVs of the pfmdr1, pfplasmepsin2 and pfgch1 genes, as well as for the molecular surveillance of the efficacy of anti-malarial drugs in field isolates. The ddPCR assays were used to detect the CNVs of the pfmdr1, pfplasmepsin2 and pfgch1 genes in field samples and validated against the results obtained with qPCR assays. A flowchart was generated including criteria for the detection of GCN with the novel ddPCR assays. The costs and time required for the ddPCR assays were discussed.
Methods
DNA samples
Development and validation of the ddPCR assays used DNA extracted from P. falciparum laboratory strains obtained from the Malaria Research and Reference Reagent Resource Center (American Type Culture Collection, Manassas, VA, USA). Parasite DNA from P. falciparum strain 3D7 (MRA-102), which carries single copies of the pfmdr1 and pfplasmespin2 genes, was used to develop and validate the ddPCR assays for the detection of the CNVs of these two genes. DNA samples of P. falciparum strain D6 originating from Sierra Leone, West Africa, which carries a single copy of the pfgch1 gene 10, were used as single copy controls for the development and validation of ddPCR assays to detect the CNVs of the pfgch1 gene. Two-fold serial dilutions of P. falciparum strain 3D7 (approximate 40,000 P. falciparum genome copies/ul based on the absolute quantification of pfβtubulin gene) and D6 (approximate 50,000 P. falciparum genome copies/ul based on the absolute quantification of pfβtubulin gene) were prepared and used to quantify the GCN, as well as to assess the accuracy and precision of the ddPCR assays.
Plasmodium falciparum strains 3D7 (isolated in Amsterdam), 7G8 (isolated in Brazil), D6 (isolated in Sierra Leone), D10 (isolated in Papua New Guinea), Dd2 (derived from cultivation), HB3 (isolated in Honduras), and W2 (isolated in Lao People’s Democratic Republic) obtained from the Malaria Research and Reference Reagent Resource Center (n = 7) were used to compare the CNVs obtained by the ddPCR and qPCR assays. In addition, the CNVs of the pfmdr1, pfplasmepsin2 and pfgch1 genes of field samples (n = 84) collected from patients with confirmed P. falciparum infections between 2015 and 2019 in Ubon Ratchathani (n = 60) and Yala (n = 24), Thailand, were determined. DNA samples were extracted using the QIAamp DNA Mini Kit (Qiagen, Hilden, North Rhine-Westphalia, Germany). DNA concentrations were measured using a Nanodrop™ spectrophotometer (Thermo Scientific, Willington, DE, USA). The study protocol was approved by the Ethical Review Committee of the Faculty of Tropical Medicine, Mahidol University (Bangkok, Thailand) (approval no. MUTM 2012-045-05).
Development of the ddPCR assays
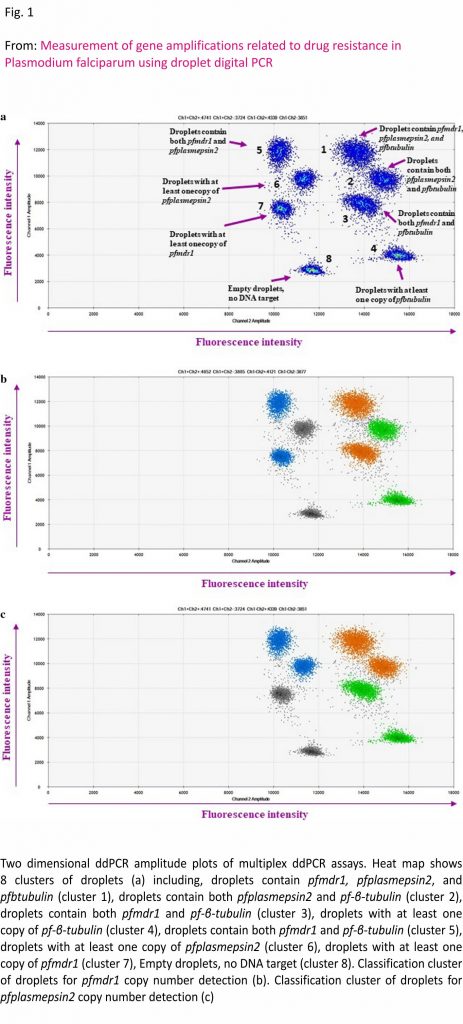
For the ddPCR assays, each 20 µL reaction contained 10 µL of ddPCR™ Supermix for Probes (Bio-Rad Laboratories, Hercules, CA, USA), 900 nM for each primer (1.8 µL of 10 µM of primer), 250 nM for each probe (0.5 µL of 10 µM probe) and 2 µL of DNA as a template. The primers and probes were previously designed for qPCR assays 6,7,11. The P. falciparum β tubulin (pfβtubulin) gene was used as the reference housekeeping gene. The ddPCR reaction was separated into 12,000– 20,000 droplets using a QX200™ Droplet Generator (Bio-Rad Laboratories) and conducted using a T100™ Thermal Cycler (Bio-Rad Laboratories). During the development of the ddPCR assays, a series of temperatures was tested to determine the optimal annealing temperature. Uniplex, duplex and multiplex ddPCR assays were developed to measure the CNVs of the pfmdr1, pfplasmepsin2 and pfgch1 genes. The optimal annealing temperature for the uniplex ddPCR assay of the pfmdr1, pfplasmepsin2, pfgch1 and pfβtubulin genes was determined to be 56 °C (Additional file 1), while that for the duplex ddPCR assay for detection of pfmdr1/pfβtubulin and pfplasmepsin2 /pfβtubulin genes was 58 °C and for the duplex ddPCR assay of the pfgch1/ pfβtubulin genes, the optimal annealing temperature was 60 °C (Additional file 2). The optimal annealing temperature for the multiplex ddPCR assay of the pfmdr1/pfplasmepsin2/pfβtubulin genes was 60 °C (Fig. 1). For validation, the ddPCR assays were performed in triplicate. After amplification, the ddPCR data were read with the use of a QX200™ Droplet Reader (Bio-Rad Laboratories) and analysed using QuantaSoft™ Software version 1.7.4 (Bio-Rad Laboratories). At least 12,000 accepted droplets were analysed 24,25. Manual thresholds were applied to distinguish between positive and negative droplets. The fluorescence intensity thresholds were determined manually for each independent experiment using the clustering tool in the QuantaSoft™ Software. Positive and negative controls were included in each run. The GCN was calculated as the ratio of the concentration (copies/µL) of the target gene to that of the reference gene.
Validation of ddPCR assays
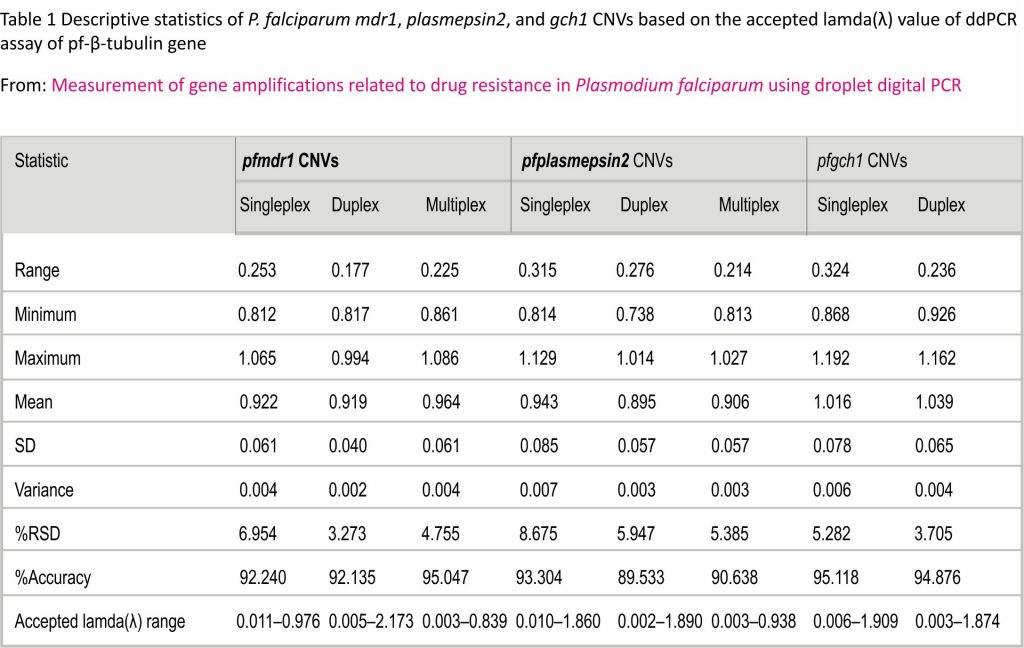
Two-fold serial dilutions of P. falciparum strain 3D7 were prepared to quantify the CNVs of the pfmdr1 and pfplasmepsin2 genes, as well as validation of the accuracy and precision of the uniplex, duplex and multiplex ddPCR assays. Two-fold serial dilutions of P. falciparum strain D6 were prepared for validation of the uniplex and duplex ddPCR assays of the CNV of the pfgch1 gene. The GCN was determined by three independent ddPCR runs. The accuracy of the ddPCR assays was calculated as % accuracy = 100 − %error and %error = the absolute difference between 1 and the GCN determined with the ddPCR assays. The precision of the ddPCR assays was calculated as the per cent relative standard deviation (%RSD) = standard deviation/average × 100. Since the DNA samples might contain both P. falciparum and human DNA, the limit of the GCN, as determined with the ddPCR assays, was quantified based on the lambda (λ) value, which was calculated as λ = ln (number of negative droplets/numbers of accepted droplets). The limit of GCN quantification of the ddPCR assays is the range of the λ value providing a %RSD value of greater than 20% and a %accuracy value of greater than 80% 25,26. In accordance with the guidelines of the Minimum Information for Publication of Quantitative Digital PCR Experiments 27, the GCN results of the P. falciparum reference strain were compared between the uniplex, duplex and multiplex ddPCR assays.
Assessment of pfmdr1, pfplasmespsin2 and pfgch1 CNVs of P. falciparum reference strains and field isolates from Thailand
DNA samples from the P. falciparum reference strain (n = 7) and the P. falciparum isolates from Thailand (n = 84) were used to evaluate the ddPCR assays. The CNV results obtained by the ddPCR assays were compared with the results of the qPCR assays, as previously described 6, 7, 11.
Statistical analysis
The GCN results of the P. falciparum reference strain, as determined with the uniplex, duplex and multiplex ddPCR assays, were compared using the independent samples median test with IBM SPSS Statistics for Windows, version 22.0 (IBM Corporation, Armonk, NY, USA). The kappa statistic was used to identify agreements between the GCN results obtained with the ddPCR assays and those obtained with the qPCR assays with the use of IBM SPSS Statistics for Windows, version 22
Results
Development and validation of ddPCR assays for CNV measurements Accuracy of the ddPCR assays
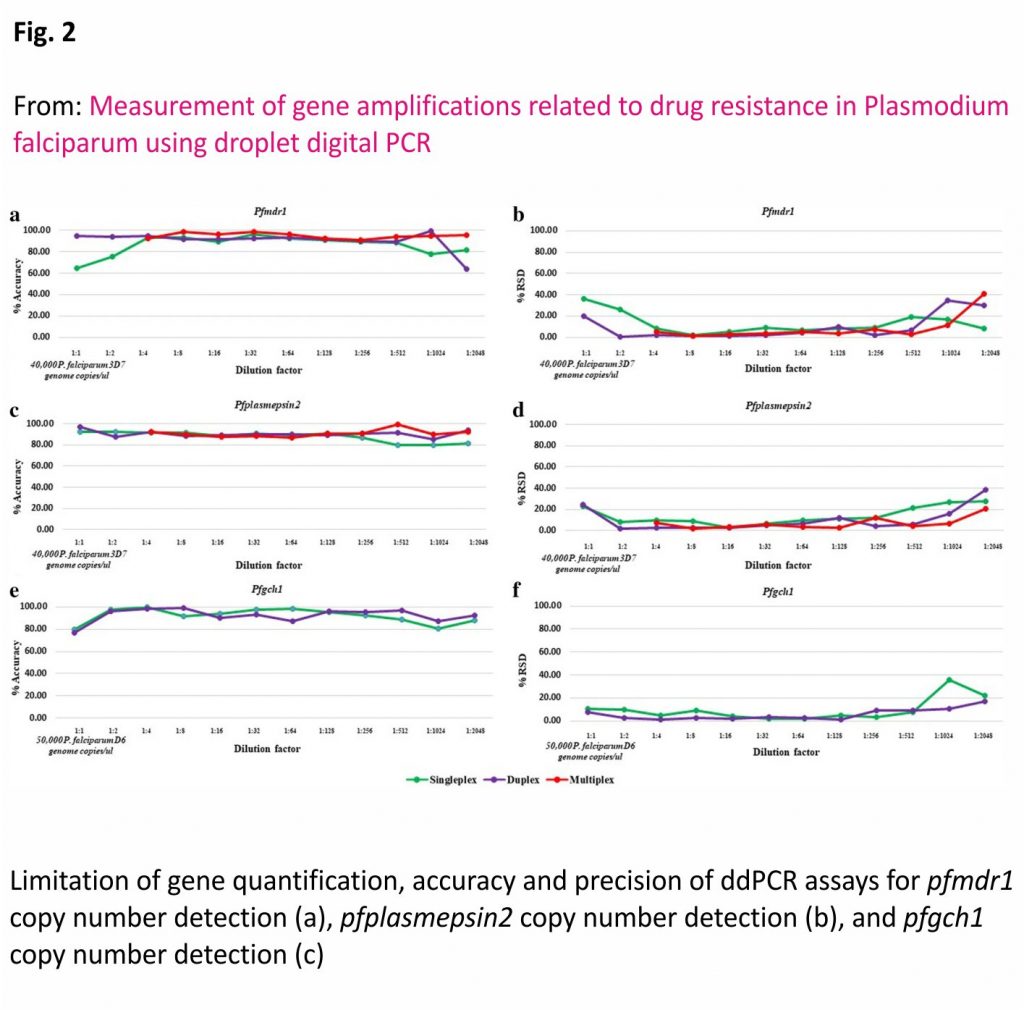
As shown in Fig. 2, the accuracies of the uniplex, duplex and multiplex ddPCR assays were 65–96%, 64–99% and 91–99%, respectively, for measurement of the pfmdr1 GCN, and 76–96%, 85–97% and 87–99%, respectively, for measurement of the pfplasmepsin2 GCN. Meanwhile, the accuracies of the uniplex and duplex ddPCR assays for measurement of the pfgch1 GCN were 80–100% and 77–99%, respectively.
The precision of ddPCR assays
As shown in Fig. 2, the %RSD values of the uniplex, duplex and multiplex ddPCR assays were 2–36%, 0–35% and 1–41 %, respectively, for detection of the pfmdr1 GCN, and 3–28%, 2–39% and 2–21%, respectively, for detection of the pfplasmepsin2 GCN. Meanwhile, the %RSD values of the uniplex and duplex ddPCR assays were 2–36% and 1–17% for detection of the pfgch1 GCN.
Limitation of GCN quantification
As shown in Table 1, the accepted range of λ values of the uniplex, duplex and multiplex ddPCR assays were 0.011– 0.987, 0.005–2.178 and 0.003–0.842, respectively, for the pfmdr1 GCN, and 0.010–1.870, 0.002–1.890 and 0.003– 0.941, respectively, for the pfplasmepsin2 GCN. The accepted range of λ values of the uniplex and duplex ddPCR assays were 0.006–1.915 and 0.003–1.877, respectively, for the pfgch1 GCN. Based on the limitation of GCN quantification, the average accuracy and %RSD value of the multiplex ddPCR assay were 95% and 5%, respectively, for measurement of the pfmdr1 GCN, and 91% and 5%, respectively, for measurement of the pfplasmepsin2 GCN. The accuracy and %RSD value of the duplex ddPCR were 95% and 4%, respectively.
Comparison between the uniplex, duplex and multiplex ddPCR assays
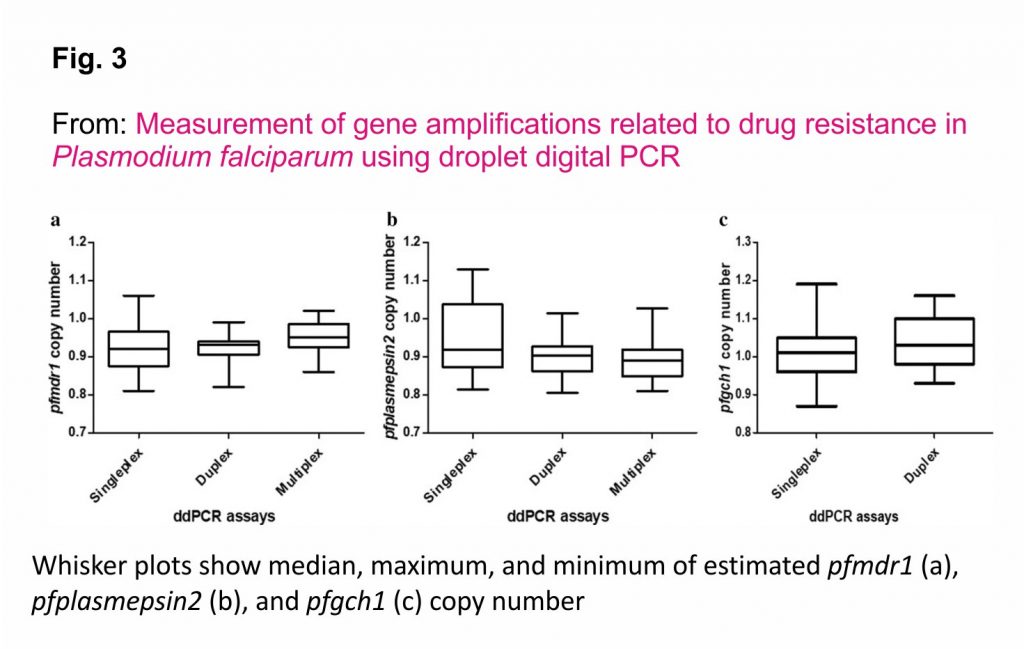
Two-fold serial dilutions of DNA from P. falciparum strain 3D7 (4, 2, 1, 0.5, 0.25, 0.125, 0.0625, 0.03125, 0.015625, 0.0078125, 0.00390625 and 0.001953125 ng/µL) were used to compare the CNVs of the pfmdr1 and pfplasmepsin2 genes obtained from the uniplex, duplex and multiplex ddPCR assays. In addition, two-fold serial dilutions of DNA from P. falciparum strain D6 (8, 4, 2, 1, 0.5, 0.25, 0.125, 0.0625, 0.03125, 0.015625, 0.0078125 and 0.00390625 ng/µL) were used for comparison of the CNVs of the pfgch1 gene obtained with the uniplex and duplex ddPCR assays. As shown in Fig. 3, there were no significant differences in the pfmdr1 (p = 0.363) and pfplasmepsin2 (p = 0.330) GCNs, as determined with the uniplex, duplex and multiplex ddPCR assays. In addition, there was no significant difference for detection of the pfgch1 GCN between the uniplex and duplex ddPCR assays (p = 0.276).
Standardized analytical workflow of ddPCR analysis for quantification of GCN
A flowchart, including validation criteria, for a standardized analytical workflow of ddPCR analysis, was designed based on the observed limit of quantification of the optimal accuracy and precision. Control samples, as well as positive and negative controls, were included for each ddPCR assay. The accepted criteria used for the ddPCR assay are that the results of the negative control are negative, those of the positive single-copy control are positive (ratio = 0.80– 1.20) and those of the positive multiple copy control are positive (ratio > 1.20). Each ddPCR reaction contained at least 12,000 droplets. The GCN results were considered acceptable at a λ value of 0.003–0.800 for quantification of the pfmdr1 and pfplasmepsin2 genes with the multiplex ddPCR assay, and 0.003– 1.900 for quantification of the pfgch1 gene with the duplex ddPCR assay. GCNs were calculated as the ratio of the concentrations (copies/µL) of the target and references genes (single GCN ratio of 0.8–1.2 and multiple GCN ratio of > 1.20).
Agreement of GCN results between the ddPCR and qPCR assays
DNA samples from the P. falciparum reference strain (n = 7) and P. falciparum strain 3D7, 7G8, D10, DD2, HB3, W2 and D6 were collected to compare the pfmdr1, pfplasmepsin2 and pfgch1 GCNs determined with the ddPCR and qPCR assays. The results showed 100% agreement between the ddPCR and qPCR assays (κ = 1) (Fig. 4, Additional file 3).
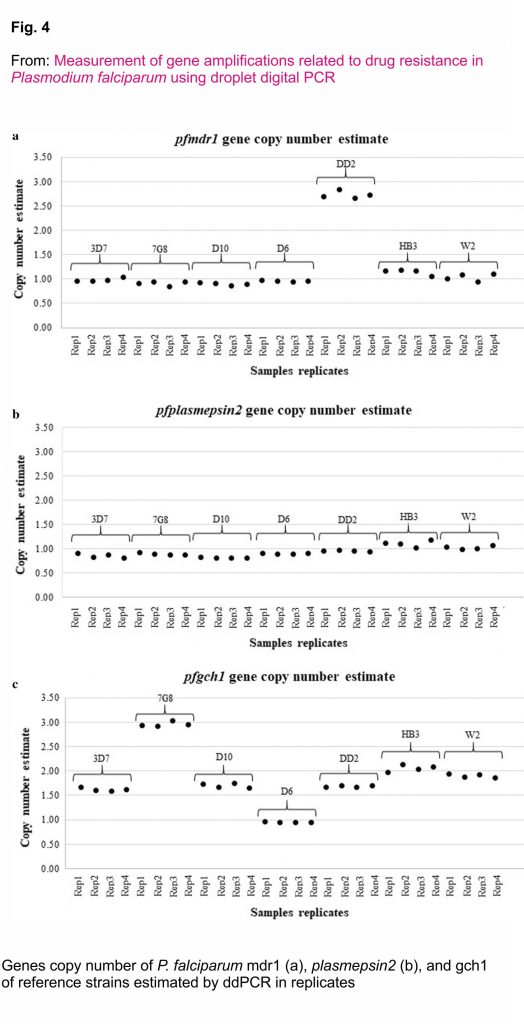 CNVs of the pfmdr1, pfplasmespsin2 and pfgch1 genes of P. falciparum isolates from Thailand
CNVs of the pfmdr1, pfplasmespsin2 and pfgch1 genes of P. falciparum isolates from Thailand
The extracted DNA samples were determined the CNVs in the pfmdr1, pfplasmespsin2 and pfgch1 genes following the standardized analytical workflow obtained from this study. The results showed that the pfmdr1 and pfplasmepsin2 GCNs were amplified in 15% and 27% of samples from Ubon Ratchathani, Northeastern Thailand, suggested evidence of selection whereas no amplification in isolates from Yala, Southern Thailand. For the pfgch1 GCN was amplified in 50% of samples from Yala, while no amplification in isolates from Ubon Ratchathani (Fig. 5, Additional file 4). Comparisons of the results of the ddPCR and qPCR assays were 100% in agreement with CNV assessments of the pfmdr1, pfplasmepsin2 and pfgch1 genes.
Cost and turn‐around time of ddPCR assays
The costs of the uniplex, duplex and multiplex ddPCR assays to determine the CNVs of the pfmdr1, pfplasmepsin2 and pfgch1 genes were 10.40, 5.50 and 5.70 USD per sample, respectively. The turn-around times for the uniplex, duplex and multiplex ddPCR assays of 96 samples were 12, 6 and 6 h, respectively.
Discussion
The ddPCR has been suggested as the reliable alternative method for high-throughput GCNs quantification 12,13. Previously, ddPCR assay was developed for screening gene deletions and duplications in human genes such as Breast cancer type 1 susceptibility protein (BRCA1) which plays a significant role in carcinogenesis of breast and ovarian cancer 20, Leucine-rich repeats and immunoglobulin-like domains 1 (LRIG1) which may be determinants of breast cancer prognostic marker 28, and mitochondrial DNA (mtDNA) which varies during ageing and disease progression 29. In malaria, qPCR assay is the most commonly used assay for the identification of genes associated with anti-malarial drug resistance 6,7,11. Here, ddPCR assays were developed and validated for accurate assessment of the CNVs of several P. falciparum resistance genes. GCNs estimated by the qPCR assay was measured based on exponential curves and calculated by the formula; (Ct of a target gene − Ct of a reference gene) of sample − (Ct of a target gene − Ct of a reference gene) of reference sample or = 2−ΔΔCt, while estimate with the ddPCR assays was measured based on the ratio of the absolute gene concentrations of the target and reference genes 13. Therefore, GCNs of ddPCR were measured without requiring the reference sample.
Although the ddPCR assay was suggested for accurate GCNs quantification, a previous study demonstrated that the accuracy and precision of GCNs quantification were reduced when using a too high or too low concentration of the target genes 30. Similarly, the results of the present study demonstrated that a higher or lower concentration of the target gene might affect the accuracy and precision of the ddPCR assays. As a consequence, optimal concentrations of the target genes are required for accurate detection of the GCNs. Here, an optimal DNA template was evaluated to accurately and precisely determine the GCN based on the λ value, which is estimated from the numbers of negative and accepted droplets generated by the ddPCR assays. So, the limit of quantification of the multiplex ddPCR assay of pfmdr1 and pfplasmepsin2 were based on the λ value which is between 0.003 and 0.8 and the limit of quantification of the duplex ddPCR assay is also based on the λ value which is between 0.003 and 1.9 for duplex ddPCR assay (Fig. 6).
To reduce the cost and turn-around time, multiplex ddPCR assays were developed for the detection of both the pfmdr1 and pfplasmepsin2 genes in a single reaction. The results showed that there were no significant differences in the GCN assessments between the assays, favouring the multiplex, rather than the uniplex, ddPCR assay as the preferred method 27. In, addition, a duplex ddPCR assay was also developed for the detection of the pfgch1 GCN instead of the uniplex ddPCR assay. Although there was no significant difference between the GCNs results obtained from the singleplex and multiplex ddPCR assay, for the culture strain experiments, the pfmdr1 and pfplasmepsin2 copy number relative to pfβtubulin ratio is consistently below 1.0 with medians near 0.9 for all suggested that the results obtained from multiplex ddPCR would be further normalized with a number to provide more accurate value.
Compared to the uniplex ddPCR assay, the use of the duplex ddPCR assay can reduce costs by 47% from 10.40 to 5.47 USD, while the estimated cost of qPCR is around 6.7 USD per sample. Duplex ddPCR assay can reduce the required assay time by 50% from 12 to 6 h. Moreover, the use of the multiplex ddPCR PCR assay to detect the pfmdr1 and pfplasmepsin2 GCNs reduced costs by 72% from 20.80 to 5.73 USD and reduced the required time by 75% from 24 to 6 h.
The prevalence of CNVs of the pfmdr1, pfplasmepsin2, which are the molecular markers of mefloquine, piperaquine resistance respectively, and pfgch1 linked to pfdhfr/pfdhps mutations which resulted in sulfadoxine-pyrimethamine resistance were assessed in Thai samples collected from the year 2015–2019. The GCN results obtained with the ddPCR and qPCR assays were in 100% agreement. The proportion of isolates with amplified pfmdr1 remains at low prevalence or zero in the two locations. This could be the results of low drug pressure as mefloquine was discontinued as a national policy for treatment of uncomplicated falciparum malaria in Thailand since 2013. Since then piperaquine has been replaced 31. The results of pfplasmepsin2 gene amplification associated with piperaquine resistance were in agreement with previous publications 32,33, which showed amplification of pfplasmepsin2 in Northeast Thailand from 2011 to 2018. This suggested piperaquine resistance in P. falciparum is prevalent in Northeast Thailand.
Although sulfadoxine-pyrimethamine (SP) anti-malarial treatment was no longer used as a national policy for the treatment of uncomplicated falciparum malaria in Thailand from 1990, the results of the present study revealed substantial amplification of the pfgch1 in Yala, near the Malaysian border. The persistence of high prevalence of antifolate resistance haplotypes in Thailand may be explained by several factors, such as continued drug pressure from non-malarial antifolate drugs such as trimethoprim and sulfamethoxazole 34, therefore, antifolate gene mutations /amplification might have been sustained because of the continued presence of this antifolate drug pressure. The prevalence of the pfmdr1, pfplasmepsin2 and pfgch1 GCNs obtained from this study might be useful for surveillance of the efficacy of anti-malarial drugs.
Conclusions
Uniplex, duplex and multiplex ddPCR assays for detection of the CNVs of the P. falciparum mdr1, plasmepsin2 and gch1 genes were developed and validated. The results confirmed the accuracy and precision of the proposed assays, which reduced the cost and turn-around time for surveillance of the efficacy of anti-malarial drugs. The assay is a valuable additional tool for genetic surveillance of anti-malarial drug resistance.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Abbreviations
DNA: Deoxyribonucleic acid
ddPCR: Droplet digital PCR
GCN: Gene copy number
PCR: Polymerase chain reaction
18S rRNA: 18s ribosomal RNA gene
pfmdr1: P. falciparum multidrug resistance 1 gene
pfgch1: P. falciparum GTP cyclohydrolase 1 gene
pfplasmepsin2 : P. falciparum plasmepsin2 gene
nM: Nanomolar
µL: Microliter
References
1. Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, Tarning J, et al. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2009;361: 455–67.
2. Phyo AP, Nkhoma S, Stepniewska K, Ashley EA, Nair S, McGready R, et al. Emergence of artemisinin-resistant malaria on the western border of Thailand: a longitudinal study. Lancet. 2012;379: 1960–6.
3. Ashley EA, Dhorda M, Fairhurst RM, Amaratunga C, Lim P, Suon S, et al. Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2014;371:411–23.
4. Denis MB, Tsuyuoka R, Poravuth Y, Narann TS, Seila S, Lim C, et al. Surveillance of the efficacy of artesunate and mefloquine combination for the treatment of uncomplicated falciparum malaria in Cambodia. Trop Med Int Health. 2006;11:1360–6.
5. Amaratunga C, Lim P, Suon S, Sreng S, Mao S, Sopha C, et al. Dihydroartemisinin-piperaquine resistance in Plasmodium falciparum malaria in Cambodia: a multisite prospective cohort study. Lancet Infect Dis. 2016;16:357–65.
6. Price RN, Uhlemann AC, Brockman A, McGready R, Ashley E, Phaipun L, et al. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet. 2004;364:438–47.
7. Witkowski B, Duru V, Khim N, Ross LS, Saintpierre B, Beghain J, et al. A surrogate marker of piperaquine-resistant Plasmodium falciparum malaria: a phenotype-genotype association study. Lancet Infect Dis. 2017;17:174–83.
8. Amato R, Lim P, Miotto O, Amaratunga C, Dek D, Pearson RD, et al. Genetic markers associated with dihydroartemisinin-piperaquine failure in Plasmodium falciparum malaria in Cambodia: a genotype-phenotype association study. Lancet Infect Dis. 2017;17:164–73.
9. Hamour S, Melaku Y, Keus K, Wambugu J, Atkin S, Montgomery J, et al. Malaria in the Nuba Mountains of Sudan: baseline genotypic resistance and efficacy of the artesunate plus sulfadoxine-pyrimethamine and artesunate plus amodiaquine combinations. Trans R Soc Trop Med Hyg. 2005;99:548–54.
10. Kidgell C, Volkman SK, Daily J, Borevitz JO, Plouffe D, Zhou Y, et al. A systematic map of genetic variation in Plasmodium falciparum. PLoS Pathog. 2006;2:e57.
11. Nair S, Miller B, Barends M, Jaidee A, Patel J, Mayxay M, et al. Adaptive copy number evolution in malaria parasites. PLoS Genet. 2008;4: e1000243.
12. Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83:8604–10.
13. Pinheiro LB, Coleman VA, Hindson CM, Herrmann J, Hindson BJ, Bhat S, et al. Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal Chem. 2012; 84:1003–11.
14. Dingle TC, Sedlak RH, Cook L, Jerome KR. Tolerance of droplet-digital PCR vs real-time quantitative PCR to inhibitory substances. Clin Chem. 2013; 59:1670–2.
15. Bhat S, Curach N, Mostyn T, Bains GS, Griffiths KR, Emslie KR. Comparison of methods for accurate quantification of DNA mass concentration with traceability to the international system of units. Anal Chem. 2010; 82:7185–92.
16. Campomenosi P, Gini E, Noonan DM, Poli A, D’Antona P, Rotolo N, et al. Imperatori A: A comparison between quantitative PCR and droplet digital PCR technologies for circulating microRNA quantification in human lung cancer. BMC Biotechnol. 2016; 16:60.
17. Vogelstein B, Kinzler KW. Digital PCR. Proc Natl Acad Sci USA. 1999;96: 9236–41.
18. Nakano M, Komatsu J, Matsuura S, Takashima K, Katsura S, Mizuno A. Single-molecule PCR using water-in-oil emulsion. J Biotechnol. 2003; 102: 117–24.
19. Ito T, Kawashima Y, Fujikawa T, Honda K, Makabe A, Kitamura K, et al. Rapid screening of copy number variations in STRC by droplet digital PCR in patients with mild-to-moderate hearing loss. Hum Genome Var. 2019;6:41.
20. Oscorbin I, Kechin A, Boyarskikh U, Filipenko M. Multiplex ddPCR assay for screening copy number variations in BRCA1 gene. Breast Cancer Res Treat. 2019;178: 545– 55.
21. Harmala SK, Butcher R, Roberts CH. Copy number variation analysis by droplet digital PCR. Methods Mol Biol. 2017;1654:135–49.
22. Koepfli C, Nguitragool W, Hofmann NE, Robinson LJ, Ome-Kaius M, Sattabongkot J, et al. Sensitive and accurate quantification of human malaria parasites using droplet digital PCR (ddPCR). Sci Rep. 2016;6:39183.
23. Srisutham S, Saralamba N, Malleret B, Renia L, Dondorp AM, Imwong M. Four human Plasmodium species quantification using droplet digital PCR. PLoS ONE. 2017;12:e017577 1.
24. Hughesman CB, Lu XJ, Liu KY, Zhu Y, Poh CF, Haynes C. A robust protocol for using multiplexed droplet digital PCR to quantify somatic copy number alterations in clinical tissue specimens. PLoS One. 2016;11: e0161274.
25. Ludlow AT, Shelton D, Wright WE, Shay JW. ddTRAP: a method for sensitive and precise quantification of telomerase activity. Methods Mol Biol. 2018;1768:513–29
26. Bhat S, Herrmann J, Armishaw P, Corbisier P, Emslie KR. Single-molecule detection in nanofluidic digital array enables accurate measurement of DNA copy number. Anal Bioanal Chem. 2009;394: 457–67.
27. Huggett JF, Foy CA, Benes V, Emslie K, Garson JA, Haynes R, et al. The digital MIQE guidelines: minimum information for publication of quantitative digital PCR experiments. Clin Chem. 2013;59:892–902.
28. Faraz M, Tellstrom A, Ardnor CE, Grankvist K, Huminiecki L, Tavelin B, et al. LRIG1 gene copy number analysis by ddPCR and correlations to clinical factors in breast cancer. BMC Cancer. 2020;20:459
29. Memon AA, Zoller B, Hedelius A, Wang X, Stenman E, Sundquist J, et al. Quantification of mitochondrial DNA copy number in suspected cancer patients by a well-optimized ddPCR method. Biomol Detect Quantif. 2017;13:32–9.
30. Deprez L, Corbisier P, Kortekaas AM, Mazoua S, Beaz Hidalgo R, Trapmann S, et al. Validation of a digital PCR method for quantification of DNA copy number concentrations by using certified reference material. Biomol Detect Quantif. 2016;9: 29–39.
31. WHO. Country cooperation strategy, Thailand: 2017–2021. Geneva, World Health Organization. 2017.
32. van der Pluijm RW, Imwong M, Chau NH, Hoa NT, Thuy-Nhien NT, Thanh NV, et al. Determinants of dihydroartemisinin-piperaquine treatment failure in Plasmodium falciparum malaria in Cambodia, Thailand, and Vietnam: a prospective clinical, pharmacological, and genetic study. Lancet Infect Dis. 2019;19: 952–61.
33. Imwong M, Dhorda M, Myo Tun K, Thu AM, Phyo AP, Proux S, et al. Molecular epidemiology of resistance to antimalarial drugs in the Greater Mekong subregion: an observational study. Lancet Infect Dis. 2020;20: 1470–80.
34. Iyer JK, Milhous WK, Cortese JF, Kublin JG, Plowe CV. Plasmodium falciparum cross-resistance between trimethoprim and pyrimethamine. Lancet. 2001;358:1066–7.
Acknowledgements
We would like to thank Ms. Jureeporn Duanguppama, Mr. Chanon Kunason, Ms. Watcharee Pagornrat, and Ms. Wanassanan Madmanee for their help.
Funding
This study was supported by Thailand Science Research and Innovation (TSRI) and National research council of Thailand, RTA6280006 and Faculty of Tropical Medicine, Mahidol University Thailand and the Wellcome Trust Mahidol University-Oxford Tropical Medicine Research Programme.
Author information
Affiliations
Department of Clinical Microscopy, Faculty of Allied Health Sciences, Chulalongkorn University, Bangkok, Thailand
Suttipat Srisutham
Mahidol-Oxford Tropical Medicine Research Unit, Faculty of Tropical Medicine, Mahidol University, Bangkok, Thailand
Kanokon Suwannasin, Arjen M. Dondorp & Mallika Imwong
Division of Vector-Borne Diseases, Department of Disease Control, Ministry of Public Health, Nonthaburi, Thailand
Rungniran Sugaram
Centre for Tropical Medicine and Global Health, Nuffield Department of Medicine, University of Oxford, Oxford, UK
Arjen M. Dondorp
Department of Molecular Tropical Medicine and Genetics, Faculty of Tropical Medicine, Mahidol University, Bangkok, Thailand
Mallika Imwong
Contributions
SS and MI contributed to study design. RS collected samples. SS and KS undertook laboratory work. SS and MI analysed data. SS, AD and MI drafted the manuscript. All authors read and approved the final manuscript.
Corresponding author
Correspondence to Mallika Imwong.
Ethics declarations
Ethics approval and consent to participate
Ethical approvals for the study were obtained from the ethical review committees of the Faculty of Tropical Medicine, Mahidol University (MUTM 2012-045-05). Informed consent was obtained from all participants.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Credits: Srisutham, S., Suwannasin, K., Sugaram, R. et al. Measurement of gene amplifications related to drug resistance in Plasmodium falciparum using droplet digital PCR. Malar J 20, 120 (2021). https://doi.org/10.1186/ s12936-021-03659-5