Moses Ikegbunam1,2*, Abone Harrison1, Chukwudi Egbuche3, Nwasolu Obidi1,2, Judith Mbamalu2, Enyi Emmanuel1, Offojebe Kosisochukwu1, Mercy Ezeunala3, Nzeukwu Chibumma4, Ifeyinwa Onochie-Igbinedion1, Joy Igwe5, Joy Nnanna6, Dorothy Ezeagwuna4, Vincent Duru4, Frances Nworji7, Charles Esimone1,2
1 Department of Pharmaceutical Microbiology and Biotechnology, Nnamdi Azikiwe University, Awka Nigeria. 2 Molecular Research foundation for students and scientists, Nnamdi Azikiwe University, Awka, Nigeria. 3 National Institute for Pharmaceutical Research and Development (NIPRD), Idu, Abuja, Nigeria. 4 Department of Parasitology and Entomology, Nnamdi Azikiwe University, Awka, Nigeria. 5 Department of Pharmaceutical microbiology and Biotechnology, Abia state University, Uturu, Abia State. 6 Department of Biological Sciences (Microbiology Unit), Dennis Osadabay University, Asaba Delta State. 7 Department of Applied Biochemistry, Nnamdi Azikiwe University, Awka, Nigeria. *email: mn.ikegbunam@unizik.edu.ng
ABSTRACT
Introduction
The genetic diversity of Plasmodium falciparum correlates with its pathogenicity, therefore, the design of evidence-based intervention strategies to eradicate malaria requires genetic diversity surveillance. This study characterised the allelic frequencies and genetic diversity of P. falciparum parasites isolated from Awka, Nigeria.
Materials and Methods
Genomic DNA was extracted from 177 P. falciparum isolates, and the polymorphic regions of the msp2 and glurp genes were genotyped by nested polymerase chain reaction (PCR).
Results
Two msp2 alleles (3D7 and FC27) were analysed. The 3D7 (93.55%) msp2 allelic family was predominant in msp2 positive isolates. Polyclonal msp2 infection was observed in 24 (38.71%) isolates. Twenty-one distinct msp2 alleles were detected, with fragment sizes ranging from 200 bp to 1200 bp. The 300 bp allelic fragment (26.83%) was predominant for the 3D7 allele, while the 400 bp allelic fragment (29.54%) was predominant for the FC27 allele. The multiplicity of infection (MoI) in msp2 was 2.03, and the expected Heterozygosity (He) was 0.34. 69 isolates (38.98%) were positive for the RII repeat region of the glurp gene. For the glurp gene, nine alleles were detected for fragment sizes ranging from 200 bp to 1150 bp, and the most prevalent allelic fragment was 200 bp (19%). The MoI and He for the glurp gene were 0.45 and 0.98, respectively.
Conclusions
The high level of polyclonal infections with P. falciparum parasites observed in this study indicates extensive genetic diversity in the study area. The data provide essential baseline information that can be implemented in developing malaria control strategies and elimination in the study area and Nigeria
INTRODUCTION
Nigeria has the highest burden of malaria globally. It is a major public health concern in Nigeria, with about 200,000 deaths from the disease annually, the primary victims being children <5 yrs and pregnant women 1. Plasmodium falciparum is the deadliest and most prevalent plasmodium species associated with malaria infection in sub-Saharan Africa 1,2. The extensive genetic diversity of P. falciparum strains poses a challenge to efforts to eradicate malaria. It possibly contributes to malaria pathology by suppressing acquired immunity and prompting the emergence of drug resistance and insecticide resistance variants 3,4,5. Therefore, genomic surveillance of the parasite is essential to develop an effective control strategy to eradicate malaria in Nigeria.
An established method to survey the parasite is by targeted genotyping of merozoite surface protein (msp-2) and glutamate-rich protein (glurp) genes as they contain polymorphic regions that are used as markers for defining genetic variation in P. falciparum malaria and determining the multiplicity of infection (MoI) 4,6,7. The msp-2 glycoprotein is an asexual blood stage antigen that consists of 5 polymorphic blocks, with block 3 being the most polymorphic. The two main allelic families of msp-2, FC27 and 3D7, are based on the polymorphic regions of the central repeat sequences 8,9. The glurp protein is an antigen expressed in both the pre-erythrocytic and erythrocytic stages of the parasite, as well as on the surface of newly released merozoites. This antigen consists of three regions, with the immunodominant C-terminal repetitive region (R2) being the most polymorphic 10. The msp-2 and glurp proteins are also considered promising candidate antigens for the development of a malaria vaccine as they are targeted by cytophilic antibodies and are associated with natural immune protection against clinical malaria 11,12.
Understanding the genetic diversity of P. falciparum in different geographical regions of Nigeria is essential for developing new and effective malaria control interventions. Currently, there is limited information on the genetic diversity and multiplicity of P. falciparum infection in southeast Nigeria. This study aimed to evaluate the allelic frequency of msp2 and glurp genes in P. falciparum parasites isolated in Awka, an urban city in southeast Nigeria. This research will contribute information on disease pathogenesis and immunity acquisition in Nigeria. It also provides information that is beneficial for the development of an effective malaria vaccine.
MATERIALS AND METHODS
This survey was conducted to assess the allelic frequency of P. falciparum msp2 and glurp genes. A total of 179 participants, aged 6 months to 68 years, were recruited between February 2019 and January 2020. Participants were included if they presented with malaria-related symptoms (e.g., fever, chills, headache) at Chukwuemeka Odumegwu Ojukwu University Teaching Hospital, Awka, Anambra State, Nigeria. The recruitment was based on clinical suspicion of malaria, and participants were screened using a rapid diagnostic test (RDT) for malaria before enrolment. Written informed consent was obtained from all participants or their legal guardians before inclusion in the study.
Study site and population
The study was conducted in Awka, Anambra State, Nigeria, a region with a tropical climate characterised by a wet season from April to October and a dry season from November to March. The area experiences an average annual rainfall of 1,200 mm and a temperature range of 25–32°C. Malaria transmission in this region is perennial, with peaks during the rainy season. Control measures include the use of insecticide-treated nets (ITNs) and intermittent preventive treatment for pregnant women (IPTp).
Participants were recruited from outpatient clinics, with inclusion criteria as follows: Age between 4 months and 60 years, presentation with clinical signs and symptoms suggestive of malaria and positive RDT for malaria before study enrolment. Exclusion criteria included participants on malaria treatment within the last two weeks or those unwilling to provide informed consent.
Sample collection, preparation and parasite detection
Venous blood samples (2 mL) were collected into EDTA tubes from each participant. RDT was performed using the SD Bioline Malaria Ag Pf/Pan (South Korea), which detects P. falciparum and non-falciparum species. Samples positive by RDT were preserved at −20°C until genomic DNA extraction. DNA extraction was performed using the Quick DNA Miniprep kit (Zymo Research, USA) following the manufacturer’s protocol.
Molecular genotyping of P. falciparum msp2 and glurp genes
The polymorphic regions of the msp2 and glurp genes were amplified in a nested PCR reaction. The primary PCR conditions for both the msp2 and glurp genes were an initial denaturation step of 95 °C for 5 min followed by 30 cycles of 95 °C for 1 min, 54 °C for 1 min, 72 °C for 1 min, and a final extension of 72 °C for 5 min. The nested PCR parameters for the glurp gene were identical to the primary reaction; only the annealing temperature was adjusted to 59 °C. For the msp2 gene, the nested PCR conditions were initial denaturation at 94°C for 5 min, followed by 30 cycles at 94 °C for 10 s, 57 °C for 30 s, and 72 °C for 40 s. The final cycle had a prolonged extension at 72 °C for 3 min. The primers targeting the RII region of glurp 13 and the 3D7 and FC27 regions of msp2 14 are shown in Table 1. The PCR products were separated on a 1.5% agarose gel, stained with ethidium bromide, and visualised using a UV transilluminator (Vilber, France).
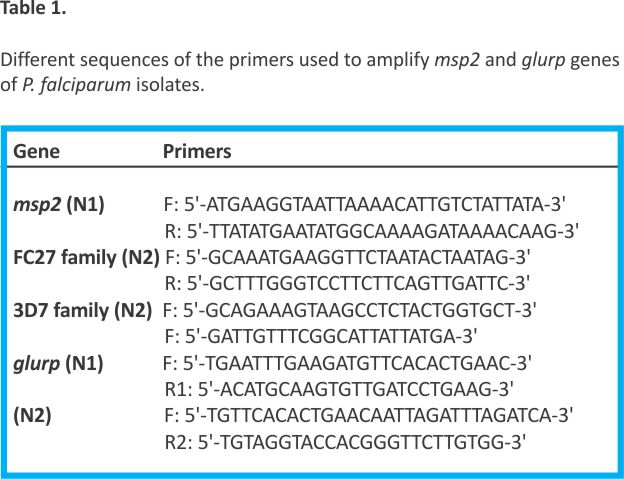 Heterozygosity and multiplicity of infection
Heterozygosity and multiplicity of infection
The expected heterozygosity index (He) was calculated using the formula:
Where n = number of isolates analysed and Pi = the frequency of the ith allele in the population. The multiplicity of infection (MoI) was calculated by dividing the total number of fragments detected in an antigenic marker by the number of samples positive for that same marker. Samples where only one genotype was detected per allelic family were monoclonal, while samples with two or more genotypes per allelic family were polyclonal P. falciparum infections.
Data analysis
The heterozygosity index (He) and multiplicity of infection (MoI) were calculated. The presence of polyclonal infections was determined based on detecting multiple alleles within the same sample.
Ethical approval
The Chukwuemeka Odumegwu Ojukwu University Teaching Hospital’s Ethics Review Board granted ethical approval for the study. (COOUTH/CMAC/ETH.C/ Vol.1/0035). Informed consent was obtained from the parent or legal guardian of each child before being included in the study.
RESULTS
The study population consisted of 53.03% female and 46.97% male patients aged 6 months to 68 years. The mean age of participants was 18.67 ± 0.32 yrs. A total of 179 blood samples were analysed for Plasmodium spp. using a malaria RDT kit, which confirmed the presence of P. falciparum in 177 (99%) samples.
Genetic diversity of P. falciparum infection
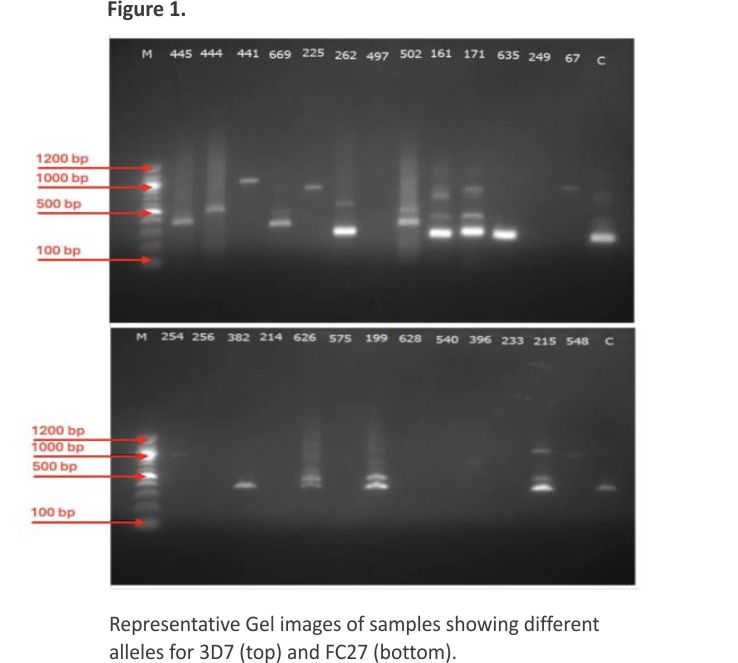
PCR amplification was successful in 35.02% (62/177) of the isolates for the msp2 gene and 38.98% (69/177) of the isolates for the glurp gene (Table 2). For msp2, the Fc27 allele had a frequency of 45.16% (28/62), while the predominant 3D7 allele had a frequency of 93.55% (58/62). Monoclonal infections were identified in 38 isolates (61.29%), with 34 isolates (54.84%) positive for the 3D7 allele and 4 isolates (6.45%) positive for the FC27 allele. Polyclonal infections (FC27+IC3D7) were detected in 24 isolates (38.71%). The RII repeat region of the glurp gene was detected in all 69 isolates. Representative gel pictures are presented in Figure 1.
 Allelic frequency of msp2 and glurp genes
Allelic frequency of msp2 and glurp genes
The allelic genotyping data revealed the polymorphic nature of P. falciparum parasites in Awka. In the msp2 and glurp genes, different allelic types were identified. Alleles of msp2 and glurp were successful in 69 isolates. Nine alleles with fragment sizes ranging from 200 to 1150 bp were identified. The most prevalent allelic fragment was 200 bp (45.24%), while both the 1050 bp and 1150 bp allelic fragments were the least prevalent (2.38%) (Figure 2)
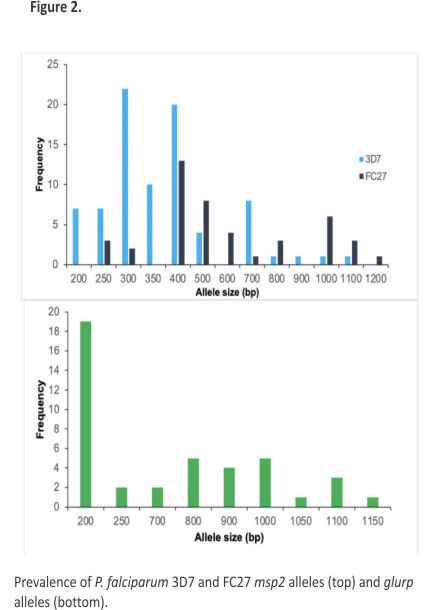 Multiplicity of infection (MoI)
Multiplicity of infection (MoI)
The MoI for the msp2 gene (MoI = 2.03) was high, as was the heterozygosity (He = 0.34). The MoI and He for the glurp gene were 0.45 and 0.98, respectively.
DISCUSSION
Nigeria is a malaria-endemic country, and an increase in the genetic diversity of P. falciparum strains could lead to more complex infections and the emergence of more virulent or drug-resistant variants, endangering efforts to eradicate the disease 15. Genetic diversity and polymorphism are key in the acquisition of anti-malaria parasite immunity 16,17. Therefore, determining the frequency of Plasmodium genotypes in different geographical locations would facilitate the development of effective control strategies. Polymorphic markers in P. falciparum isolates were used to examine the genetic diversity and complexity of parasite populations in patients with symptomatic malaria at the Chukwuemeka Odumegwu Ojukwu Teaching Hospital, Awka, Nigeria.
In this study, allele-specific genotyping of msp2 in P. falciparum isolates reveals high allelic diversity in Awka, Nigeria. The high malaria transmission rate, the incidence of mixed illnesses, and the subsequent exposure of locals to mosquito bites may be responsible for this trend in the research area. For msp2, the 3D7 alleles were predominant, with a 93.54% occurrence. This data is consistent with studies from Kaduna 18, Ibadan 19, Anambra 7, and north-central Nigeria 20,21 that reported a high prevalence of the 3D7 family, in symptomatic patients, but the results contrast with a study by Ojurongbe et al. 22 in Osogbo, Nigeria, which reported the FC27 allele as the predominant allele. The result is in agreement with other studies conducted in sub-Saharan Africa 23,24, South America 16, and Asia 17,25. The results suggest that the frequency of 3D7 alleles strongly correlates with symptomatic malaria. The result is conflicting as the 3D7 allele is associated with asymptomatic malaria infections, and it is thought to offer protection against clinical disease 26. The contrasting observation from various studies indicates a need to understand the influence of human genetic factors on the antigenicity of msp2 alleles as the variations observed in different studies may be attributed to immune selection pressures.
Among the msp2 positive P. falciparum isolates, polyclonal infections were observed in 38.71% of participants. Complex infections marked by multiclonality impact drug efficacy, severity of disease and the population diversity of the parasite 4. Polyclonal infections are associated with low levels of protective malaria antibodies, increased prevalence of drug-resistant parasites and possibility of recrudescence 27,28. More importantly, malaria vaccine efficacy studies on the msp2 gene have shown that vaccination with only one msp2 variant induces an allele-specific response. In one study involving a vaccine that comprised the 3D7 allelic family, vaccinated patients showed an increase in morbidity associated with the FC27 alleles 29. Subsequent studies have shown that chimeric vaccines 30 and vaccines containing both msp2 allelic families 31 induce a strain-transcending immune response. This necessitates that the malaria vaccine design incorporates both allelic variants of the msp2 gene to account for the increase in polyclonal infections observed in this study.
The high genetic diversity for msp2 is consistent with observations in malaria-endemic regions 4. The high genetic diversity may correlate with the rate of polyclonal infections and the intensity of malaria transmission in the region 4. The multiplicity of P. falciparum infections (MoI) for msp2 reported in this study is similar to studies from Nnewi 7 and Ibadan 32 in Nigeria. However, other studies from southwest Nigeria 33,34,35 and Pahang, Malaysia 36 reported a lower MoI, while a study on children living close to a lake in Taabo, Côte d’Ivoire, reported a higher MoI. The multiplicity of infection (MoI), i.e., the number of different P. falciparum strains co-infecting a single host in many malaria-endemic areas is a common feature and has been reported to vary with age, parasite density, immune status, epidemiological settings, and transmission intensity 37. The MoI observed in this study could indicate high transmission levels of the parasite.
The RII region of the glurp was also shown to have a high prevalence in the study population, with a 38.98% occurrence. The study by Ullah et al. 6 showed a higher prevalence of 70% in Pakistan, consistent with another study conducted in southwestern Nigeria 32. Furthermore, a study in Osogbo, Nigeria, reported that the genetic diversity of the R2 polymorphic region of the glurp gene remained diverse despite the implementation of the artemisinin-based combination therapy ACT therapy in the study area 38. The glurp gene is an antigen of P. falciparum that is highly conserved, present in all stages of the malaria parasite and associated with clinical immunity. These attributes make it a promising biomarker for diagnosis and the development of vaccines against malaria.
CONCLUSIONS
The present study shows that there is a high level of polyclonal P. falciparum infections in the population. The P. falciparum parasites harbour multiple gene alleles with high MoI. This indicates the extensive genetic diversity of P. falciparum infection in the study area. The data provides important baseline information to guide malaria control and elimination strategies in the study area and across Nigeria.
ACKNOWLEDGEMENTS
We would like to express our sincere gratitude to the staff of COUTH for their assistance with sample collection, the dedicated participants who made this study possible, the staff of the Molecular Research Foundation for their invaluable support, and my colleagues for their insightful feedback.
COMPETING INTERESTS
The author declares no competing interests.
REFERENCES
1. World Health Organization: World malaria report 2023. Geneva, World Health Organization. https://tinyurl.com/4h4jd9nx
2. Alemayehu A: Biology and epidemiology of Plasmodium falciparum and Plasmodium vivax gametocyte carriage: implication for malaria control and elimination. Parasite Epidemiol. Control 2023, 21:e0 0295. Doi: 10.1016/j.parepi. 2023.e00295
3. Abukari Z, Okonu R, Nyarko SB, Aminata C Lo et al.: The diversity, multiplicity of infection and population structure of P. falciparum parasites circulating in asymptomatic carriers living in high and low malaria transmission settings of Ghana. Genes 2019, 10:434. Doi: 10.3390/genes 10060434
4. Opute AO, Akinkunmi JA, Funsho AO, Obaniyi AK et al.: Genetic diversity of Plasmodium falciparum isolates in Nigeria. A review. Egypt. J. Med. Hum. Genet. 2022, 23:129. Doi: 10.1186/s43 042-022-00340-7
5. Ahouidi A, Ali M, Almagro-Garcia J, Amambua-Ngwa A et al.: An open dataset of Plasmodium falciparum genome variation in 7,000 worldwide samples. Wellcome Open Res. 2021, 6:42. Doi: 10.12688/wellcome openres.16168.2
6. Ullah I, Khan A, Israr M, Shah M et al.: Genomic miscellany and allelic frequencies of Plasmodium falciparum msp-1, msp-2 and glurp in parasite isolates. PLoS One 2022, 17:e0264654. Doi: 10.1371/journal.pone.0264654
7. Ikegbunam MN, Anagu LO, Duru C, Nworu CS et al.: Genetic diversity and allelic frequency of antigenic markers in Plasmodium falciparum isolates from Nnewi district in Nigeria. J. Infect. Dev. Ctries 2022, 16:557–563. Doi: 10.3855/jidc.14815
8. Beeson JG, Drew DR, Boyle MJ, Feng G et al.: Merozoite surface proteins in red blood cell invasion, immunity and vaccines against malaria. FEMS Microbiol. Rev. 2016, 40:343–72. Doi: 10.1093/ femsre/fuw001
9. Mohammed H, Assefa A, Chernet M, Wuletaw Y et al.: Genetic polymorphisms of Plasmodium falciparum isolates from Melka-Werer, North East Ethiopia based on the merozoite surface protein-2 (msp-2) gene as a molecular marker. Malar. J. 2021, 20:85. Doi: 10.1186/s129 36-02103625-1
10. Kaur H, Sehgal R, Goyal K, Makkar N et al.: Genetic diversity of Plasmodium falciparum merozoite surface protein-1 (block 2), glutamate-rich protein and sexual stage antigen Pfs25 from Chandigarh, North India. Trop. Med. Int. Health 2017, 22:1590-1598. Doi: 10.1111/tmi.12990
11. Milet J, Sabbagh A, Migot-Nabias F, Luty AJ et al.: Genome-wide association study of antibody responses to Plasmodium falciparum candidate vaccine antigens. Genes Immun. 2016, 17:110-117. Doi: 10.1038/gene. 2015.59
12. Fall AK, Kana IH, Dechavanne C, Garcia-Senosiain A et al.: Naturally acquired antibodies from Beninese infants promote Plasmodium falciparum merozoite-phagocytosis by human blood leukocytes: implications for control of asymptomatic malaria infections. Malar. J.2022,21: 356. Doi: 10.1186/s12936 -022- 04361-w
13. Nguetse CN, Ojo JA, Nchotebah C, Ikegbunam MN et al.: Genetic Diversity of the Plasmodium falciparum glutamate-rich protein R2 region before and twelve years after introduction of artemisinin combination therapies among febrile children in Nigeria. Am. J. Trop. Med. Hyg. 2018, 98: 667–676. Doi: 10.4269/ajtmh. 17-0621
14. Snounou G, Zhu X, Siripoon N, Jara W et al.: Biased distribution of msp1 and msp2 allelic variants in Plasmodium falciparum populations in Thailand. Trans. R. Soc. Trop. Med. Hyg. 1999, 93:369–374. Doi: 10.1016/ s0035-9203(99)90120-7
15. Oyedeji SI, Awobode HO, Anumudu C, Kun J et al..: Genetic diversity of Plasmodium falciparum isolates from naturally infected children in north-central Nigeria using the merozoite surface protein-2 as molecular marker. Asian Pac. J. Trop. Med. 2013, 6:589–594. Doi: 10.1016/s19957645 (13)60102-9
16. Chenet SM, Branch OH, Escalante AA, Lucas CM et al.: Genetic diversity of vaccine candidate antigens in Plasmodium falciparum isolates from the Amazon basin of Peru. Malar. J. 2008, 7:93. Doi: 10.1186/1475-2875-7-93
17. Zakeri S, Bereczky S, Naimi P, Gill JP et al..: Multiple genotypes of the merozoite surface proteins 1 and 2 in Plasmodium falciparum infections in a hypoendemic area in Iran. Trop. Med. Int. Health 2005, 10:1060–1064. Doi: 10.1111/j.1365-3156.2005. 01477.x
18. Dikwa KB, Yahaya UA, Maikaje DB, Suleiman AB: Genetic diversity of Plasmodium falciparum isolated from symptomatic and asymptomatic individuals in parts of Kaduna metropolis, Kaduna state, Nigeria. Am. J. Microbiol. Res. 2020, 8:83–92. Doi: 10.12 691/ajmr-8-3-2
19. Amodu OK, Olaniyan SA, Omotade OO: Changes in Plasmodium falciparum population dynamics in two populations at different periods in Ibadan, South-west Nigeria. Afr. J. Biomed. Res. 2015, 18:17–22. https://tinyurl. com/5f5299px
20. Oyedeji SI, Awobode HO, Kun J: Limited genetic diversity and low multiplicity of Plasmodium falciparum infections in Children with severe malaria in Lafia, North-central Nigeria. J. Exp. Clin. Med. 2013, 5:143–147. Doi: 10.1016/ j.jecm.2013.06.014
21. Oyedeji SI, Bassi PU, Oyedeji SA, Ojurongbe O et al..: Genetic diversity and complexity of Plasmodium falciparum infections in the microenvironment among siblings of the same household in North-Central Nigeria. Malar. J. 2020, 19:338. Doi: 10.1186/s12936-020-03415-1
22. Ojurongbe O, Fagbenro-Beyioku AF, Adeyeba OA, Kun JF: Allelic diversity of merozoite surface protein 2 gene of P falciparum among children in Osogbo, Nigeria. West Indian Med. J. 2011, 60:19–23. https://tinyurl. com/38386zfp
23. Takala S, Branch O, Escalante AA, Kariuki S et al.: Evidence for intragenic recombination in Plasmodium falciparum: identification of a novel allele family in block 2 of merozoite surface protein-1: Asembo Bay Area Cohort Project XIV. Mol. Biochem. Parasitol. 2002, 125: 163–171. Doi: 10.1016/s0166-6851(02)00237-2
24 Mayengue PI, Ndounga M, Malonga FV, Bitemo L et al.: Genetic polymorphism of merozoite surface protein-1 and merozoite surface protein-2 in Plasmodium falciparum isolates from Brazzaville, Republic of Congo. Malar. J. 2011, 10:276. Doi: 10.1186/1475-2875-10-276
25. Yuan L, Zhao H, Wu L, Li X et al.: Plasmodium falciparum populations from northeastern Myanmar display high levels of genetic diversity at multiple antigenic loci. Acta Trop. 2013, 125:53–59. Doi: 10.1016/ j.actatropica.2012.09.008
26. al-Yaman F, Genton B, Reeder JC, Anders RF et al.: Reduced risk of clinical malaria in children infected with multiple clones of Plasmodium falciparum in a highly endemic area: a prospective community study. Trans. R. Soc. Trop. Med. Hyg. 1997, 91:602 –605. Doi: 10.1016/s0035-9203(97)90046-8
27. Biabi MF, Fogang B, Essangui E, Maloba F et al.: High Prevalence of polyclonal Plasmodium falciparum infections and association with poor IgG antibody responses in a hyper-endemic area in Cameroon. Trop. Med. Infect. Dis. 2023, 8:390. Doi: 10.3390/tropical med8080390
28. Lopez L, Koepfli C: Systematic review of Plasmodium falciparum and Plasmodium vivax polyclonal infections: Impact of prevalence, study population characteristics, and laboratory procedures. PLoS One 2021, 16:e0249382. Doi: 10.1371/journal.pone. 0249382
29. Flück C, Smith T, Beck H-P, Irion A et al..: Strain-specific humoral response to a polymorphic malaria vaccine. Infect. Immun. 2004, 72:6300–6305. Doi: 10.1128/iai.72.11.6300-6305. 2004
30. McCarthy JS, Marjason J, Elliott S, Fahey P et al.: A phase 1 trial of MSP2-C1, a blood-stage malaria vaccine containing 2 isoforms of MSP2 formulated with Montanide® ISA 720. PLoS One 2011, 6:e24413. Doi: 10. 1371/journal.pone.0024413
31. Krishnarjuna B, Andrew D, MacRaild CA, Morales RA et al.: Strain-transcending immune response generated by chimeras of the malaria vaccine candidate merozoite surface protein 2. Sci. Rep. 2016, 6:20613. Doi: 10.10 38/srep20613
32. Funwei RI, Thomas BN, Falade CO, Ojurongbe O: Extensive diversity in the allelic frequency of Plasmodium falciparum merozoite surface proteins and glutamate-rich protein in rural and urban settings of southwestern Nigeria. Malar. J. 2018, 17:1. Doi:10.1186/s12936-017-2149-5
33. Kolawole OM, Mokuolu OA, Olukosi YA, Oloyede TO: Population genomics diversity of Plasmodium falciparum in malaria patients attending Okelele Health Centre, Okelele, Ilorin, Kwara State, Nigeria. Afr. Health Sci. 2016, 16:704–711. Doi: 10.4314/ahs. v16i3.10
34. Oyebola MK, Idowu ET, Olukosi YA, Olukosi YA et al..: Genetic diversity and complexity of Plasmodium falciparum infections in Lagos, Nigeria. Asian Pac. J. Trop. Biomed. 2014, 4:S87–S91. Doi: 10.12980/apjtb.4.2014 c1301
35. Olasehinde GI, Yah CS, Singh R, Ojurongbe O et al..: Genetic diversity of Plasmodium falciparum field isolates from south western Nigeria. Afr. Health Sci. 2012, 12:355361. Doi: 10.4314/ahs. v12i3.17
36. Atroosh WM, Al-Mekhlafi HM, Mahdy MA, Saif-Ali R et al.: Genetic diversity of Plasmodium falciparum isolates from Pahang, Malaysia based on MSP-1 and MSP-2 genes. Parasit. Vectors 2011,4:233. Doi: 10.1186/1756-3305-4-233
37. Abamecha A, El-Abid H, Yilma D, Addis W et al..: Genetic diversity and genotype multiplicity of Plasmodium falciparum infection in patients with uncomplicated malaria in Chewaka district, Ethiopia. Malar. J. 2020, 19:203. Doi: 10.1186/s12936-020-03278-6
38. Nguetse CN, Ojo JA, Nchotebah C, Ikegbunam MN et al.: Genetic Diversity of the Plasmodium falciparum glutamate-rich protein R2 region before and twelve years after introduction of artemisinin combination therapies among febrile children in Nigeria. Am. J. Trop. Med. Hyg. 2018, 98: 667–676. Doi: 10.4269/ajtmh. 17-0621
Credits: Ikegbunam M, Harrison A, Egbuche C, Obidi N, Mbamalu J, Emmanuel E, Kosisochukwu O, Ezeunala M, Chibumma N, Onochie-Igbinedion I, Igwe J, Nnanna J, Ezeagwuna D, Duru V, Nworji F, Esimone C. Allelic frequency of msp2 and glurp genes in Plasmodium falciparum isolates from Awka, Anambra, Nigeria. Malariaworld J. 2025 Feb 18;16:4. doi: 10.5281/ zenodo.14886922. PMID:4006 6288; PMCID: PMC11892418.